6X3C
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6X3J
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F0224 (46) | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8YRH
 
 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | Authors: | Wang, Q.S, Li, Q.H. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
8I4U
 
 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4V
 
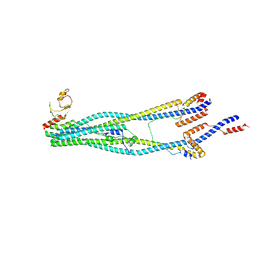 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I13
 
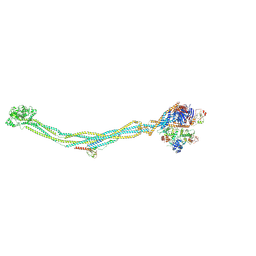 | | Cryo-EM structure of 6-subunit Smc5/6 | | Descriptor: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
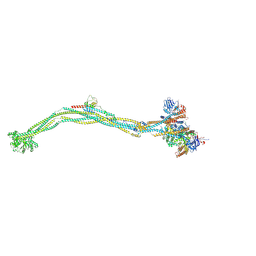 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4W
 
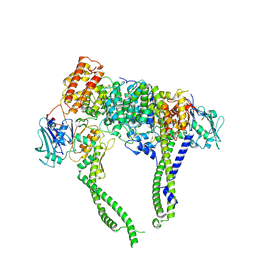 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7WMJ
 
 | | A novel chemical derivative(71) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(phenylcarbonyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMO
 
 | | A novel chemical derivative(92) of THRB agonist | | Descriptor: | 2-[[1-ethoxy-7-[4-(3-fluoranyl-5-methoxy-phenyl)carbonyl-2,6-dimethyl-phenoxy]-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMH
 
 | | A novel chemical derivative(56) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenylphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMG
 
 | | A novel chemical derivative(52) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenoxyphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMN
 
 | | A novel chemical derivative(89) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(3-methylphenyl)carbonyl-phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WML
 
 | | A novel chemical derivative(85) of THRB agonist | | Descriptor: | 2-[[7-[4-(3,5-dimethylphenyl)carbonyl-2,6-dimethyl-phenoxy]-1-ethoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WLX
 
 | | A novel chemical derivative(53) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-[4-(phenylmethyl)phenoxy]isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
6PCR
 
 | | E. coli 50S ribosome bound to compound 40o | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl (2-bromopyridin-4-yl)carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PC5
 
 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PCS
 
 | | E. coli 50S ribosome bound to compound 40e | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl [4-(trifluoromethyl)phenyl]carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PCT
 
 | | E. coli 50S ribosome bound to compound 41q | | Descriptor: | (2S)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PCQ
 
 | | E. coli 50S ribosome bound to VM2 | | Descriptor: | (3R,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-3-(propan-2-yl)-8,9,14,15,24,25,26,26a-octahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,16,22(4H,17H)-tetrone, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PC6
 
 | | E. coli 50S ribosome bound to compound 47 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6PCH
 
 | | E. coli 50S ribosome bound to compound 21 | | Descriptor: | (3R,4R,5E,10E,12E,14S,26aR)-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-8,9,14,15,24,25,26,26a-octahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,16,22(4H,17H)-tetrone, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
