7FCS
 
 | |
8V38
 
 | |
5XI1
 
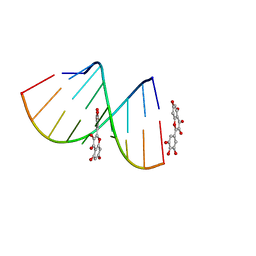 | | Structural Insight of Flavonoids binding to CAG repeat RNA that causes Huntington's Disease (HD) and Spinocerebellar Ataxia (SCAs) | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, RNA (5'-R(P*CP*CP*GP*CP*AP*GP*CP*GP*G)-3') | | Authors: | Tawani, A, Mishra, S.K, Khan, E, Kumar, A. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Myricetin Reduces Toxic Level of CAG Repeats RNA in Huntington's Disease (HD) and Spino Cerebellar Ataxia (SCAs).
ACS Chem. Biol., 13, 2018
|
|
4JHA
 
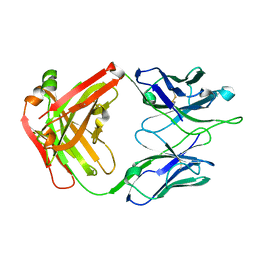 | | Crystal Structure of RSV-Neutralizing Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
4JHW
 
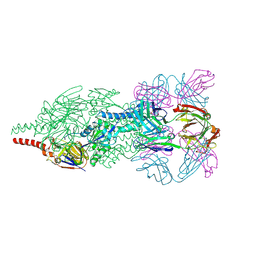 | | Crystal Structure of Respiratory Syncytial Virus Fusion Glycoprotein Stabilized in the Prefusion Conformation by Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain, Fusion glycoprotein F0 | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
2G15
 
 | | Structural Characterization of autoinhibited c-Met kinase | | Descriptor: | activated met oncogene | | Authors: | Wang, W, Marimuthu, A, Tsai, J, Kumar, A, Krupka, H.I, Zhang, C, Powell, B, Suzuki, Y, Nguyen, H, Tabrizizad, M, Luu, C, West, B.L. | | Deposit date: | 2006-02-13 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4P6D
 
 | | Structure of ribB complexed with PO4 ion | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6P
 
 | | Structure of ribB complexed with inhibitor (4PEH) and metal ions | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ZINC ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8J
 
 | | Structure of ribB | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8E
 
 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P77
 
 | | Structure of ribB complexed with substrate Ru5P | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, RIBULOSE-5-PHOSPHATE | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6C
 
 | | Structure of ribB complexed with inhibitor 4PEH | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
5Z47
 
 | | Crystal structure of pyrrolidone carboxylate peptidase I with disordered loop A from Deinococcus radiodurans R1 | | Descriptor: | DIMETHYL SULFOXIDE, Pyrrolidone-carboxylate peptidase | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of pyrrolidone-carboxylate peptidase I from Deinococcus radiodurans reveal the mechanism of L-pyroglutamate recognition.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2L94
 
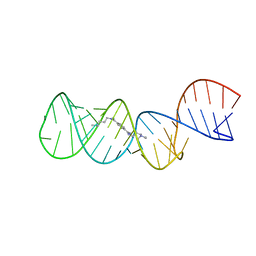 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
5Z48
 
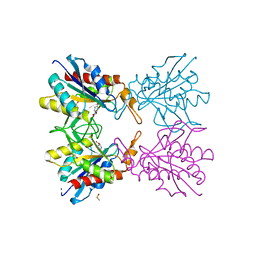 | | Crystal structure of pyrrolidone carboxylate peptidase I from Deinococcus radiodurans R1 bound to pyroglutamate | | Descriptor: | DIMETHYL SULFOXIDE, PYROGLUTAMIC ACID, Pyrrolidone-carboxylate peptidase, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structures of pyrrolidone-carboxylate peptidase I from Deinococcus radiodurans reveal the mechanism of L-pyroglutamate recognition.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3S5C
 
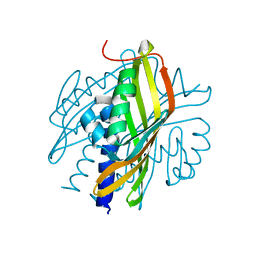 | | Crystal Structure of a Hexachlorocyclohexane dehydrochlorinase (LinA) Type2 | | Descriptor: | LinA | | Authors: | Kukshal, V, Macwan, A.S, Kumar, A, Ramachandran, R. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the hexachlorocyclohexane dehydrochlorinase (LinA-type2): mutational analysis, thermostability and enantioselectivity
Plos One, 7, 2012
|
|
6A8Z
 
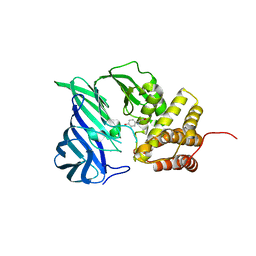 | | Crystal structure of M1 zinc metallopeptidase from Deinococcus radiodurans | | Descriptor: | SODIUM ION, TYROSINE, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Makde, R.D. | | Deposit date: | 2018-07-11 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
5XYL
 
 | |
5CAD
 
 | | Crystal structure of the vicilin from Solanum melongena revealed existence of different anionic ligands in structurally similar pockets | | Descriptor: | ACETATE ION, MAGNESIUM ION, PYROGLUTAMIC ACID, ... | | Authors: | Jain, A, Kumar, A, Salunke, D.M. | | Deposit date: | 2015-06-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the vicilin from Solanum melongena reveals existence of different anionic ligands in structurally similar pockets
Sci Rep, 6, 2016
|
|
5YZM
 
 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6A4R
 
 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | Descriptor: | ASPARTIC ACID, Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
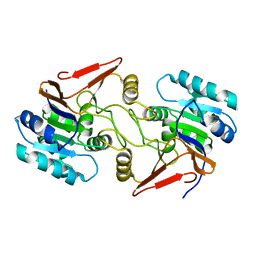 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A9U
 
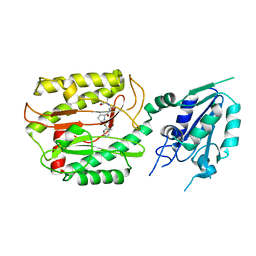 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
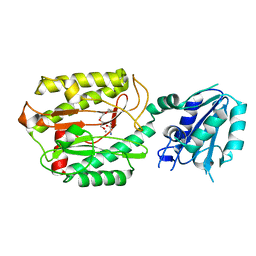 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A4T
 
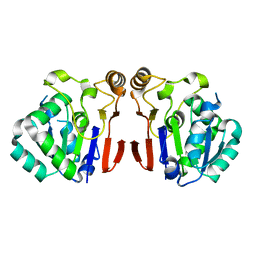 | | Crystal structure of Peptidase E from Deinococcus radiodurans R1 | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Goyal, V.G, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic triad heterogeneity in S51 peptidase family: Structural basis for functional variability.
Proteins, 87, 2019
|
|
