6KKL
 
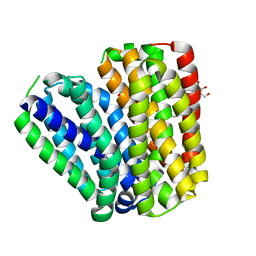 | | Crystal structure of Drug:Proton Antiporter-1 (DHA1) Family SotB, in the inward conformation (H115N mutant) | | Descriptor: | Sugar efflux transporter, nonyl beta-D-glucopyranoside | | Authors: | Xiao, Q.J, Sun, B, Zuo, Y.X, Guo, L, He, J.H, Deng, D. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Visualizing the nonlinear changes of a drug-proton antiporter from inward-open to occluded state.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
4UEB
 
 | |
2XSL
 
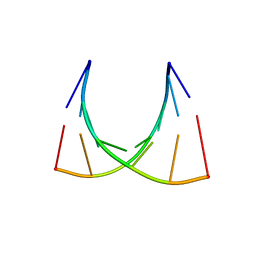 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
4OAL
 
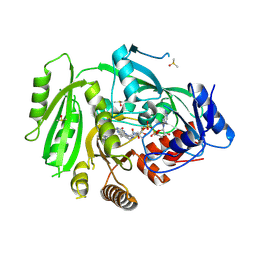 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU in alternative spacegroup | | Descriptor: | 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
4O95
 
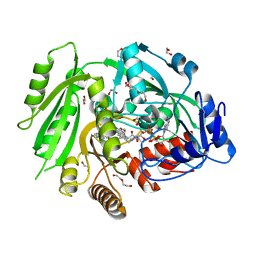 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-01 | | Release date: | 2015-04-01 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
8DNT
 
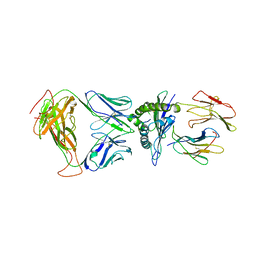 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
4UAG
 
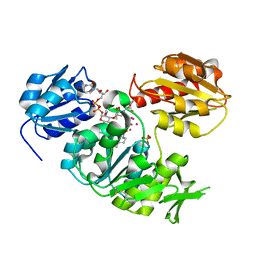 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
8E1E
 
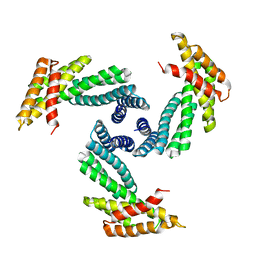 | |
4UE9
 
 | |
8ELC
 
 | | Human JNK2 bound to covalent inhibitor YL2056 | | Descriptor: | 4-(dimethylamino)-N-{4-[(3S)-3-({4-[(8R)-2-phenylpyrazolo[1,5-a]pyridin-3-yl]pyrimidin-2-yl}amino)pyrrolidine-1-carbonyl]phenyl}butanamide, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
4UEA
 
 | |
4UEC
 
 | | Complex of D. melanogaster eIF4E with eIF4G and cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4G, ... | | Authors: | Peter, D, Weichenrieder, O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Architecture of 4E-BP Translational Inhibitors Bound to Eif4E.
Mol.Cell, 57, 2015
|
|
4UE8
 
 | |
1VT7
 
 | |
1VKQ
 
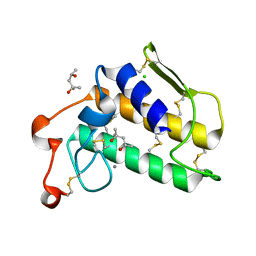 | | A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Yamane, T, Dauter, M, Dauter, Z. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VZM
 
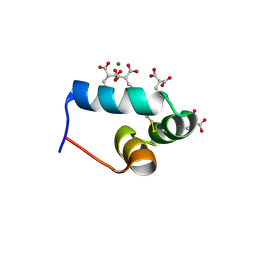 | | OSTEOCALCIN FROM FISH ARGYROSOMUS REGIUS | | Descriptor: | MAGNESIUM ION, OSTEOCALCIN | | Authors: | Frazao, C, Simes, D.C, Coelho, R, Alves, D, Williamson, M.K, Price, P.A, Cancela, M.L, Carrondo, M.A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-09-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications
Biochemistry, 44, 2005
|
|
1VPU
 
 | |
1VIP
 
 | | ANTICOAGULANT CLASS II PHOSPHOLIPASE A2 FROM THE VENOM OF VIPERA RUSSELLI RUSSELLI | | Descriptor: | PHOSPHOLIPASE A2, SULFATE ION | | Authors: | Carredano, E, Westerlund, B, Persson, B, Saarinen, M, Ramaswamy, S, Eaker, D, Eklund, H. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structures of two toxins from snake venom throw light on the anticoagulant and neurotoxic sites of phospholipase A2.
Toxicon, 36, 1998
|
|
1WAL
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) MUTANT (M219A)FROM THERMUS THERMOPHILUS | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1VJK
 
 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
1VRZ
 
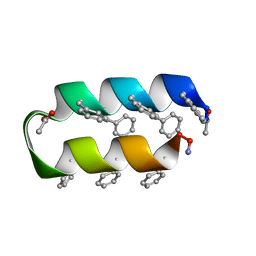 | | Helix turn helix motif | | Descriptor: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | Authors: | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
1VYQ
 
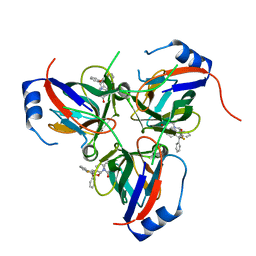 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
1W56
 
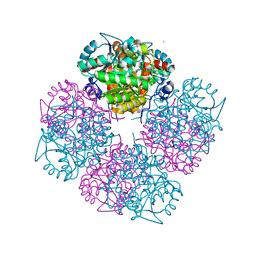 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C and D131C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W76
 
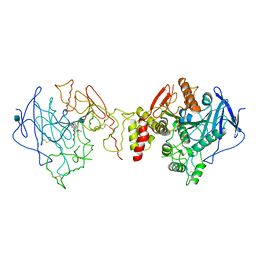 | | Orthorhombic form of Torpedo californica acetylcholinesterase (AChE) complexed with bis-acting galanthamine derivative | | Descriptor: | (-)-GALANTHAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Greenblatt, H.M, Guillou, C, Guenard, D, Badet, B, Thal, C, Silman, I, Sussman, J.L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design.
J.Am.Chem.Soc., 126, 2004
|
|
1VSV
 
 | |
