3ICO
 
 | |
3PXX
 
 | |
3IFT
 
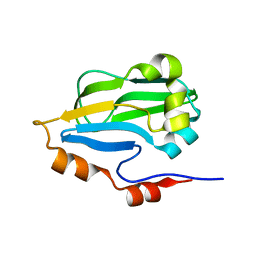 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
3QI6
 
 | |
3F0A
 
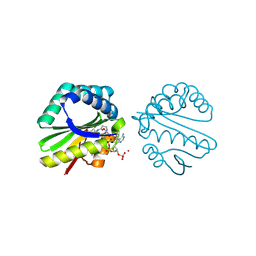 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
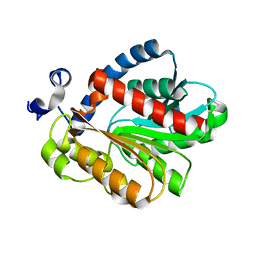
3QH4
 
 | |
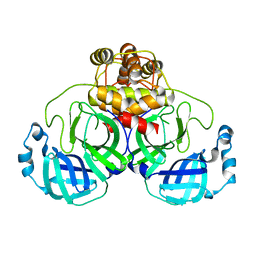
3F9G
 
 | |
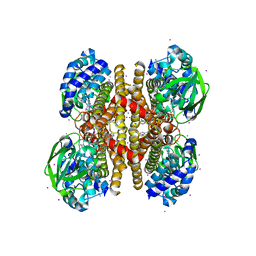
3PFD
 
 | |
3PGX
 
 | |
3QHX
 
 | |
3H81
 
 | |
3H7F
 
 | |
3S55
 
 | |
7F45
 
 | | Structure of an Anti-CRISPR protein | | Descriptor: | AcrIF5 | | Authors: | Feng, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
8GS1
 
 | | Crystal structure of AziU2-U3 complex from Streptomyces sahachiroi NRRL2485 | | Descriptor: | Azi28, Azi29, FORMIC ACID, ... | | Authors: | Cheng, Y, Li, P, Liu, W, Fang, P. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
7WRJ
 
 | |
4B9D
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
8HK0
 
 | | Crystal structure of Fic32-33 complex from Streptomyces ficellus NRRL 8067 | | Descriptor: | Acyl-CoA dehydrogenase, Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cheng, Y, Qiao, H, Liu, W, Fang, P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
7EJ2
 
 | | human voltage-gated potassium channel KV1.3 H451N mutant | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
7EJ1
 
 | | human voltage-gated potassium channel KV1.3 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4AGU
 
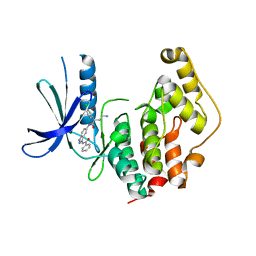 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL1 KINASE DOMAIN | | Descriptor: | CYCLIN-DEPENDENT KINASE-LIKE 1, N-(5-{[(2S)-4-amino-2-(3-chlorophenyl)butanoyl]amino}-1H-indazol-3-yl)benzamide | | Authors: | Canning, P, Sharpe, T.D, Allerston, C, Savitsky, P, Pike, A.C.W, Muniz, J.R.C, Chaikuad, A, Kuo, K, Burgess-Brown, N, Ayinampudi, V, Zhang, Y, Thangaratnarajah, C, Ugochukwu, E, Vollmar, M, Krojer, T, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Knapp, S, Bullock, A. | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
7Y04
 
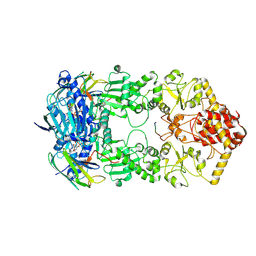 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7F83
 
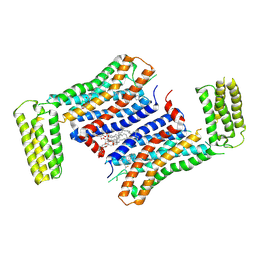 | | Crystal Structure of a receptor in Complex with inverse agonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)-1-[2-[(1R)-5-(6-methylpyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]-2,7-diazaspiro[3.5]nonan-7-yl]ethanone, Growth hormone secretagogue receptor type 1,Soluble cytochrome b562 | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
3ZKJ
 
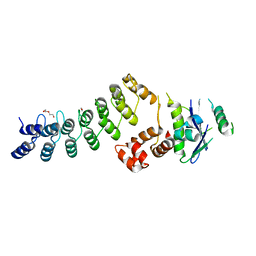 | | Crystal Structure of Ankyrin Repeat and Socs Box-Containing Protein 9 (Asb9) in Complex with Elonginb and Elonginc | | Descriptor: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Guo, K, Zhang, Y, Ayinampudi, V, Savitsky, P, Keates, T, Filippakopoulos, P, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, von Delft, F, Knapp, S, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2013-01-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
