8UPA
 
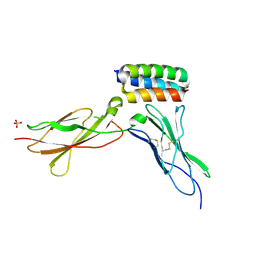 | | Structure of gp130 in complex with a de novo designed IL-6 mimetic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, De novo designed IL-6 mimetic, Interleukin-6 receptor subunit beta, ... | | Authors: | Borowska, M.T, Jude, K.M, Garcia, K.C. | | Deposit date: | 2023-10-22 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo design of miniprotein antagonists of cytokine storm inducers.
Nat Commun, 15, 2024
|
|
7VVV
 
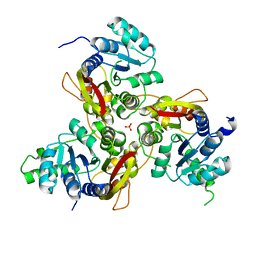 | | Crystal structure of MmtN | | Descriptor: | PHOSPHATE ION, SAM-dependent methyltransferase | | Authors: | Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVX
 
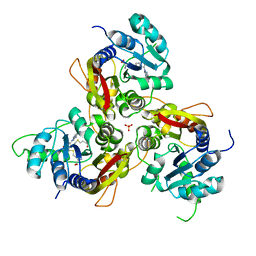 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVW
 
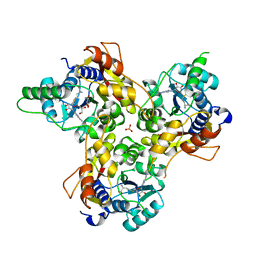 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
5D8T
 
 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
3JXB
 
 | |
5IKK
 
 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
4FPD
 
 | | Deprotonation of D96 in bacteriorhodopsin opens the proton uptake pathway | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, CHLORIDE ION, ... | | Authors: | Wang, T, Sessions, A.O, Lunde, C.S, Rouani, S, Glaeser, R.M, Facciotti, M.T, Duan, Y. | | Deposit date: | 2012-06-22 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Deprotonation of d96 in bacteriorhodopsin opens the proton uptake pathway.
Structure, 21, 2013
|
|
7XWX
 
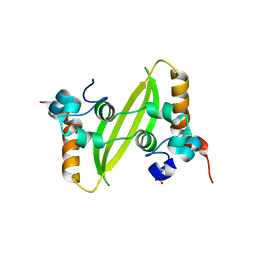 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWZ
 
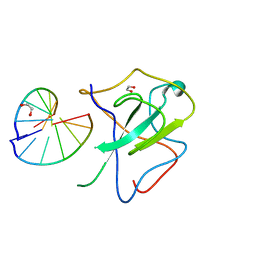 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
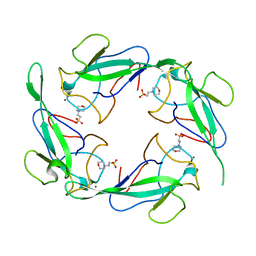 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
3JXD
 
 | |
7E0W
 
 | | Crystal Structure of BCH domain from S. pombe | | Descriptor: | Putative Rho GTPase-activating protein C1565.02c, TETRAETHYLENE GLYCOL | | Authors: | Chichili, V.P.R, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel intertwined anti-parallel dimeric structure of scaffold BCH domain regulates RhoA and RhoGAP functions
Proc.Natl.Acad.Sci.USA, 2021
|
|
5IQY
 
 | | Structure of apo-Dehydroascorbate Reductase from Pennisetum Glaucum phased by Iodide-SAD method | | Descriptor: | Dehydroascorbate reductase, IODIDE ION | | Authors: | Das, B.K, Kumar, A, Manidola, P, Arockiasamy, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase
Biochem.Biophys.Res.Commun., 473, 2016
|
|
3JXC
 
 | |
8RJX
 
 | |
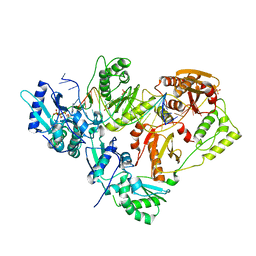
3KK1
 
 | |
8OV5
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE, 100K | | Descriptor: | CHLOROPHYLL A, PERIDININ, Peridinin-chlorophyll a-binding protein 1, ... | | Authors: | Hofmann, E, Johanning, S. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and spectroscopic characterization of the peridinin-chlorophyll a-protein (PCP) complex from Heterocapsa pygmaea (HPPCP).
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
1IJC
 
 | | Solution Structure of Bucandin, a Neurotoxin from the Venom of the Malayan Krait | | Descriptor: | bucandin | | Authors: | Torres, A.M, Kini, R.M, Nirthanan, S, Kuchel, P.W. | | Deposit date: | 2001-04-25 | | Release date: | 2001-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bucandin, a neurotoxin from the venom of the Malayan krait (Bungarus candidus).
Biochem.J., 360, 2001
|
|
7N21
 
 | | NMR structure of AnIB-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N22
 
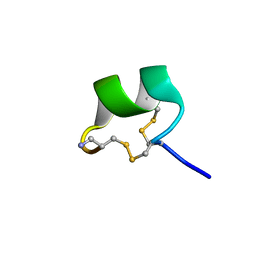 | | NMR structure of AnIB[Y(SO3)16Y]-NH2 | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
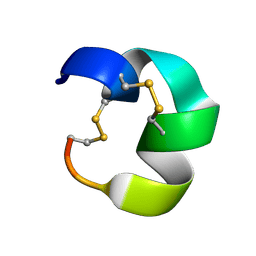 | | NMR structure of AnIB[Y(SO3)16Y]-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N0T
 
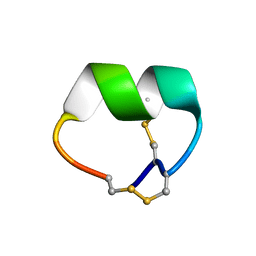 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N20
 
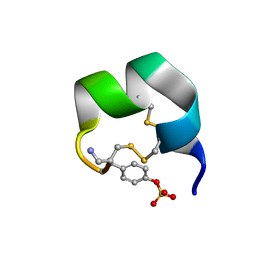 | | NMR structure of native AnIB | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7W7F
 
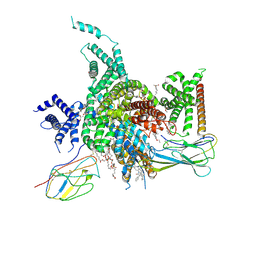 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
