5TZX
 
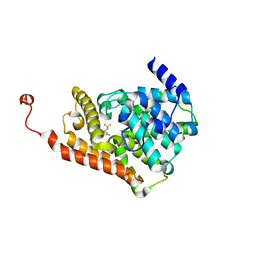 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 1-[(3-chloro-4-fluorophenyl)carbonyl]-3,3-difluoro-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | (3-chloro-4-fluorophenyl)[(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl]methanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZC
 
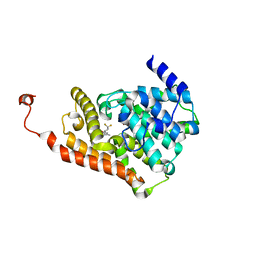 | | Crystal Structure of human PDE2a in complex with (5S)-1-[(3-bromo-4-fluorophenyl)carbonyl]-3,3-difluoro-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | (3-bromo-4-fluorophenyl)[(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl]methanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZH
 
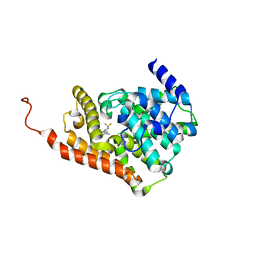 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 3,3-difluoro-1-[(4-fluorophenyl)carbonyl]-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](4-fluorophenyl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZW
 
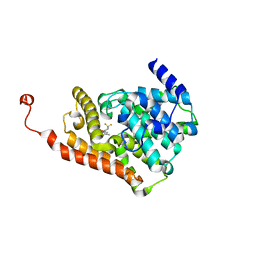 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 1-[(3,4-difluorophenyl)carbonyl]-3,3-difluoro-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](3,4-difluorophenyl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZA
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A WITH 3-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl-1-[(naphthalene-2-yl)carbonyl]piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(3S)-3-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](naphthalen-2-yl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5TZZ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 1-[(3-bromo-4-fluorophenyl)carbonyl]-3,3-difluoro-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | (3-bromo-4-fluorophenyl)[(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl]methanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
8XEP
 
 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | Descriptor: | SULFATE ION, Type IV effector MavL, Ubiquitin | | Authors: | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8I71
 
 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
8HRU
 
 | | Cryo-EM structure of ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HRI
 
 | |
8HRK
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 complex | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HRM
 
 | | Cryo-EM structure of streptavidin | | Descriptor: | Streptavidin | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HRN
 
 | | Cryo-EM structure of ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HRJ
 
 | |
8HVU
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVV
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVW
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVY
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVZ
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
4XU5
 
 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU4
 
 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU6
 
 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
8KHU
 
 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
