6ZIH
 
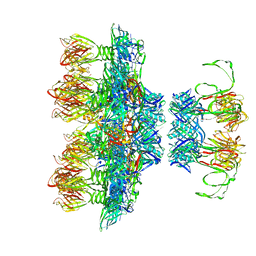 | |
6ZJJ
 
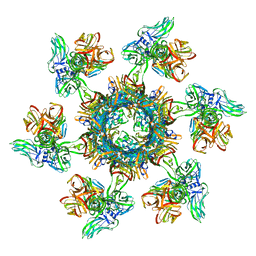 | |
4JEI
 
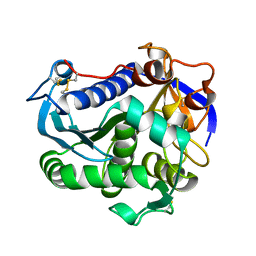 | | Nonglycosylated Yarrowia lipolytica LIP2 lipase | | Descriptor: | Lipase 2 | | Authors: | Aloulou, A, Benarouche, A, Puccinelli, D, Spinelli, S, Cavalier, J.-F, Cambillau, C, Carriere, F. | | Deposit date: | 2013-02-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of non-glycosylated Yarrowia lipolytica LIP2 lipase
To be published
|
|
4GQK
 
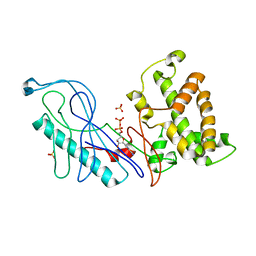 | | Structure of Native VgrG1-ACD with ADP (no cations) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, VgrG protein | | Authors: | Nguyen, V.S, Spinelli, S, Derrez, E, Durand, E, Cambillau, C. | | Deposit date: | 2012-08-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of Native VgrG1-ACD with ADP (no cations)
To be Published
|
|
2H36
 
 | |
1FWX
 
 | | CRYSTAL STRUCTURE OF NITROUS OXIDE REDUCTASE FROM P. DENITRIFICANS | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Djinovic-Carugo, K, Haltia, T, Cabrito, I, Saraste, M, Moura, J.J, Moura, I, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revisiting the Catalytic CuZ Cluster of Nitrous Oxide (N2O) Reductase. Evidence of a Bridging Inorganic Sulfur
J.Biol.Chem., 275, 2000
|
|
4HH6
 
 | | Peptide from EAEC T6SS Sci1 SciI protein | | Descriptor: | Peptide from EAEC T6SS Sci1 SciI protein, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
4HH5
 
 | | N-terminal domain (1-163) of ClpV1 ATPase from E.coli EAEC Sci1 T6SS. | | Descriptor: | BROMIDE ION, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
1LK5
 
 | | Structure of the D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, D-Ribose-5-Phosphate Isomerase, SODIUM ION | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A hyperthermostable D-ribose-5-phosphate isomerase from Pyrococcus horikoshii characterization and three-dimensional structure.
Structure, 10, 2002
|
|
1LK7
 
 | | Structure of D-Ribose-5-Phosphate Isomerase from in complex with phospho-erythronic acid | | Descriptor: | CHLORIDE ION, D-4-PHOSPHOERYTHRONIC ACID, D-Ribose-5-Phosphate Isomerase, ... | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Hyperthermostable D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii Characterization and Three-Dimensional Structure
STRUCTURE, 10, 2002
|
|
4PS2
 
 | | Structure of the C-terminal fragment (87-165) of E.coli EAEC TssB molecule | | Descriptor: | CHLORIDE ION, Putative type VI secretion protein, ZINC ION | | Authors: | Douzi, B, Logger, L, Spinelli, S, Blangy, S, Cambillau, C, Cascales, E. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the tube-sheath interface within the Type VI secretion system tail
To be Published
|
|
4PSD
 
 | | Structure of Trichoderma reesei cutinase native form. | | Descriptor: | Carbohydrate esterase family 5 | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
4RGG
 
 | |
4PSC
 
 | | Structure of cutinase from Trichoderma reesei in its native form. | | Descriptor: | Carbohydrate esterase family 5, GLYCEROL | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
7BNW
 
 | |
7BNP
 
 | |
7BBA
 
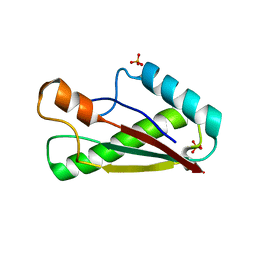 | |
4PSE
 
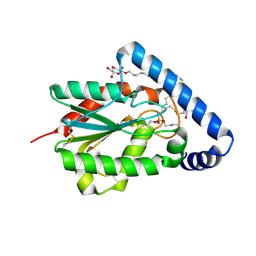 | | Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor | | Descriptor: | Carbohydrate esterase family 5, UNDECYL-PHOSPHINIC ACID BUTYL ESTER, octyl beta-D-glucopyranoside | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
3S0F
 
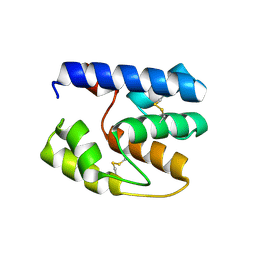 | | Apis mellifera OBP14 native apo, crystal form 2 | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
1OKS
 
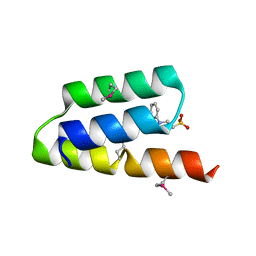 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
1P28
 
 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae in complex with a component of the pheromonal blend: 3-hydroxy-butan-2-one. | | Descriptor: | R,3-HYDROXYBUTAN-2-ONE, S,3-HYDROXYBUTAN-2-ONE, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1ORG
 
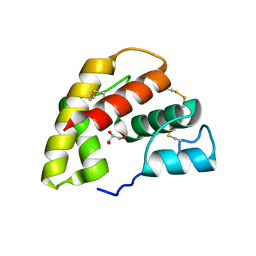 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae reveals a new mechanism of pheromone binding | | Descriptor: | GLYCEROL, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
3S0D
 
 | | Apis mellifera OBP 14 in complex with the citrus odorant citralva (3,7-dimethylocta-2,6-dienenitrile) | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dienenitrile, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
