1RXW
 
 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXZ
 
 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RWZ
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1SCE
 
 | |
1US8
 
 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1SZX
 
 | | Role Of Hydrogen Bonding In The Active Site Of Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Greenleaf, W.B, Perry, J.J, Hearn, A.S, Cabelli, D.E, Lepock, J.R, Stroupe, M.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of hydrogen bonding in the active site of human manganese superoxide dismutase.
Biochemistry, 43, 2004
|
|
1T6U
 
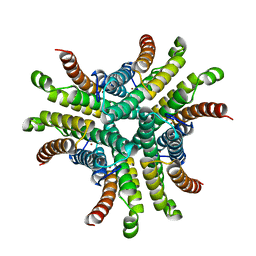 | | Nickel Superoxide Dismutase (NiSOD) Native 1.30 A Structure | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-07 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
3E67
 
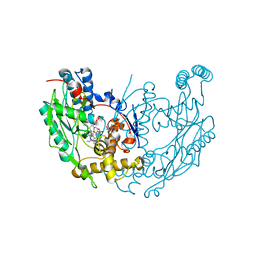 | | Murine inos dimer with inhibitor 4-MAP bound | | Descriptor: | 4-METHYLPYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E6T
 
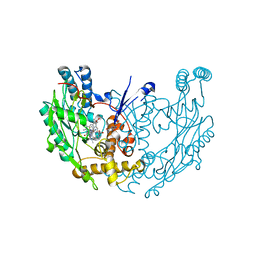 | | Structure of murine INOS oxygenase domain with inhibitor AR-C118901 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-(4'-AMINO-1'-ETHYL-5',8'-DIFLUORO-1'H-SPIRO[PIPERIDINE-4,2'-QUINAZOLINE]-1-YLCARBONYL)PICOLINONITRILE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E68
 
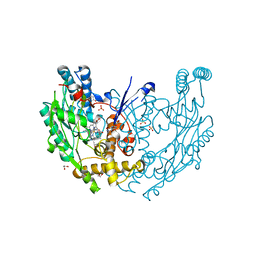 | | Structure of murine INOS oxygenase domain with inhibitor AR-C130232 | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, N-[2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL]-4-CYANOBENZAMIDE, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7M
 
 | | Structure of murine iNOS oxygenase domain with inhibitor AR-C95791 | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7S
 
 | | Structure of bovine eNOS oxygenase domain with inhibitor AR-C95791 | | Descriptor: | ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, HEME C, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7G
 
 | | Structure of human INOSOX with inhibitor AR-C95791 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3CRV
 
 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3DSC
 
 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3FY4
 
 | | (6-4) Photolyase Crystal Structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-4 photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2009-01-21 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3E7I
 
 | | Structure of murine inos oxygenase domain with inhibitor AR-C94864 | | Descriptor: | (2R)-5-FLUORO-2-(2-THIENYL)-1,2-DIHYDROQUINAZOLIN-4-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7T
 
 | | Structure of murine iNOS oxygenase domain with inhibitor AR-C102222 | | Descriptor: | 1-(6-CYANO-3-PYRIDYLCARBONYL)-5',8'-DIFLUOROSPIRO[PIPERIDINE-4,2'(1'H)-QUINAZOLINE]-4'-AMINE, 7,8-DIHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E6L
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C132283 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-CHLOROPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EAI
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | 4-({4-[(4-methoxypyridin-2-yl)amino]piperidin-1-yl}carbonyl)benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3ENM
 
 | | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 6, GLYCEROL, ... | | Authors: | Min, X, Akella, R, He, H, Humphreys, J.M, Tsutakawa, S, Lee, S.-J, Tainer, J.A, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer
Structure, 17, 2009
|
|
3DSD
 
 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
