1QQR
 
 | | CRYSTAL STRUCTURE OF STREPTOKINASE DOMAIN B | | Descriptor: | STREPTOKINASE DOMAIN B | | Authors: | Spraggon, G, Zhang, X.X, Ponting, C.P, Fox, V.F, Phillips, C, Smith, R.A.G, Jones, E.Y, Dobson, C, Stuart, D.I. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptokinse Domain B
To be Published
|
|
5V00
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (Formate condition) | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Liu, Y, Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas.
Proteins, 85, 2017
|
|
8JEL
 
 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form I | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEQ
 
 | | Crystal structure of Tiragolumab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEP
 
 | | Crystal structure of Ociperlimab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
 | | Crystal structure of TIGIT in complexed with Tiragolumab | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEN
 
 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form II | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
6LWA
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWF
 
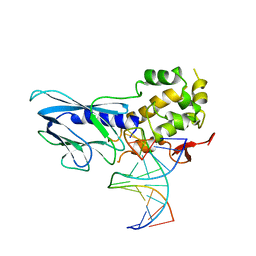 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWC
 
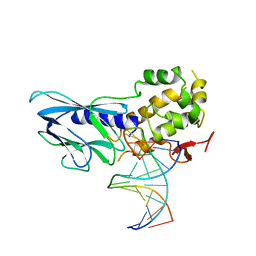 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWG
 
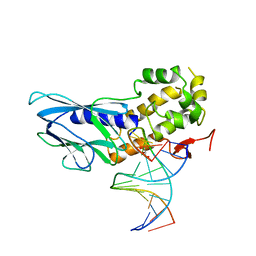 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWB
 
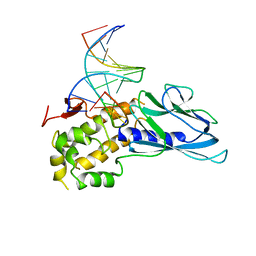 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWN
 
 | | Crystal structure of human NEIL1(R242, G249P) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWO
 
 | | Crystal structure of human NEIL1(R242, Y244H) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWM
 
 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWI
 
 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing dihydrothymine (DHT) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(TDH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWJ
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing dihydrouracil (DHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(UDH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWK
 
 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing dihydrouracil (DHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(UDH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWD
 
 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWP
 
 | | Crystal structure of human NEIL1(R242, Y244R) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWH
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing dihydrothymine (DHT) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(TDH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWL
 
 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWQ
 
 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing a C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*TP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWR
 
 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing a cleaved C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*(PDA))-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
7W3T
 
 | | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
