1AMS
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLUTARIC ACID | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
1AMR
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
1AMQ
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
2YRR
 
 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
2YRI
 
 | | Crystal structure of alanine-pyruvate aminotransferase with 2-methylserine | | Descriptor: | (S,E)-3-HYDROXY-2-((3-HYDROXY-2-METHYL-5-(PHOSPHONOOXYMETHYL)PYRIDIN-4-YL)METHYLENEAMINO)-2-METHYLPROPANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase, ... | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | hypothetical alanine aminotransferase (TTHA0173) from Thermus thermophilus HB8
To be Published
|
|
1AK7
 
 | | DESTRIN, NMR, 20 STRUCTURES | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK6
 
 | | DESTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
5XDF
 
 | |
1UDY
 
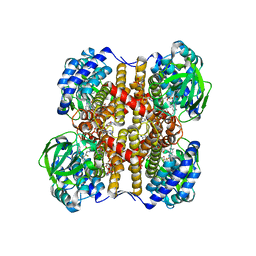 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
5BJ4
 
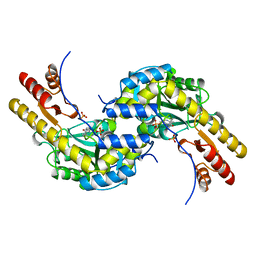 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1KA9
 
 | |
1KH3
 
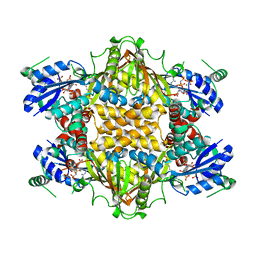 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitor | | Descriptor: | ARGININE, ASPARTIC ACID, Argininosuccinate Synthetase, ... | | Authors: | goto, m, Hirotsu, k, miyahara, i, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-11-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Argininosuccinate Synthetase in Enzyme-ATP Substrates and Enzyme-AMP Product Forms: STEREOCHEMISTRY OF THE CATALYTIC REACTION
J.Biol.Chem., 278, 2003
|
|
2DKJ
 
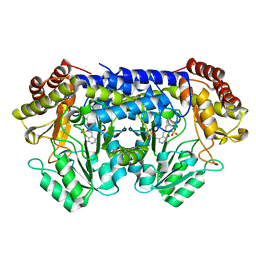 | | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, serine hydroxymethyltransferase | | Authors: | Kai, K, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase
To be Published
|
|
2DDH
 
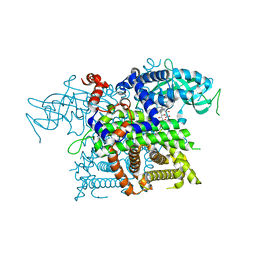 | | Crystal Structure of Acyl-CoA oxidase complexed with 3-OH-dodecanoate | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Acyl-CoA oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Keiji, T, Nakajima, Y, Miyahara, I, Hirotsu, K. | | Deposit date: | 2006-01-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Three-Dimensional Structure of Rat-Liver Acyl-CoA Oxidase in Complex with a Fatty Acid: Insights into Substrate-Recognition and Reactivity toward Molecular Oxygen.
J.Biochem.(Tokyo), 139, 2006
|
|
2DOU
 
 | | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, probable N-succinyldiaminopimelate aminotransferase | | Authors: | Omi, R, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-03 | | Release date: | 2006-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8
To be published
|
|
2DQ4
 
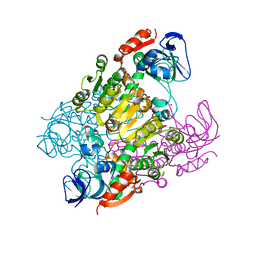 | | Crystal structure of threonine 3-dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-threonine 3-dehydrogenase, ZINC ION | | Authors: | Omi, R, Yao, T, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of threonine 3-dehydrogenase
To be Published
|
|
5BJ3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 1 | | Descriptor: | PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
8GUH
 
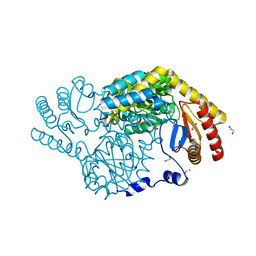 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Tris | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [4-[[[2-(hydroxymethyl)-1,3-bis(oxidanyl)propan-2-yl]amino]methyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Sphingobacterium multivorum serine palmitoyltransferase complexed with tris(hydroxymethyl)aminomethane.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8H20
 
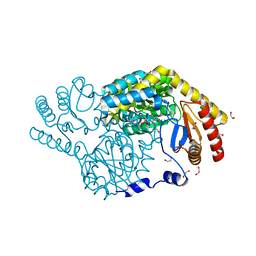 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H21
 
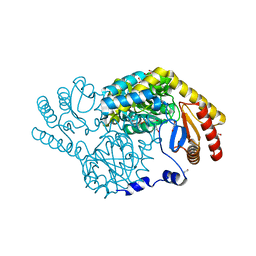 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-alanine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H29
 
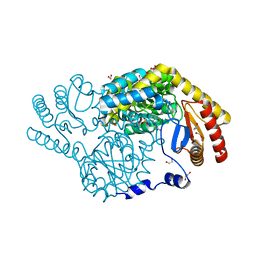 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-threonine | | Descriptor: | 1,2-ETHANEDIOL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1W
 
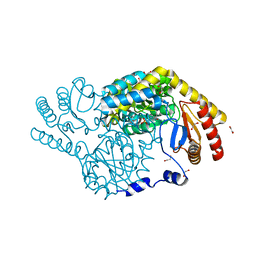 | | Serine Palmitoyltransferase from Sphingobacterium multivorum | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Q
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-serine | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Y
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-homoserine | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
6A0R
 
 | | Homoserine dehydrogenase from Thermus thermophilus HB8 unliganded form | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
