7XPM
 
 | | Ancestral ADH WT | | Descriptor: | 1,2-ETHANEDIOL, A64 | | Authors: | Chen, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2022-05-04 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a versatile ancestral ADH with high activity and thermostability
To Be Published
|
|
7YII
 
 | | Carboxylesterase - RoCE | | Descriptor: | Non-heme haloperoxidase | | Authors: | Dou, Z, Jia, P, Ni, Y, Xu, G.C. | | Deposit date: | 2022-07-16 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Carboxylesterase - RoCE
To Be Published
|
|
7YMU
 
 | | Structure of Alcohol dehydrogenase from [Candida] glabrata | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sun, Z.W, Liu, Y.F, Xu, G.C, Ni, Y. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationla design of CgADH from candida glarata for asymmetric reduction of azacycolne.
To Be Published
|
|
7YMB
 
 | |
8J44
 
 | | Reductive Aminase RA34-WT | | Descriptor: | 6-phosphogluconate dehydrogenase NAD-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
8J43
 
 | | Reductive aminase RA29-WT | | Descriptor: | 1,2-ETHANEDIOL, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
7C1E
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Y127W) | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, Y.F, Zhou, J.Y, Liu, Y.F, Xu, G.C, Ni, Y. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering an Alcohol Dehydrogenase from Kluyveromyces polyspora for Efficient Synthesis of Ibrutinib Intermediate
Adv.Synth.Catal., 2021
|
|
5ZED
 
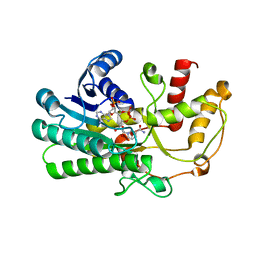 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5Z2X
 
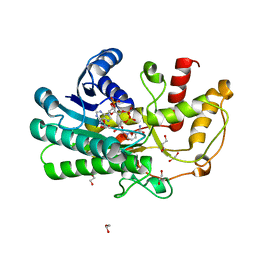 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5ZEC
 
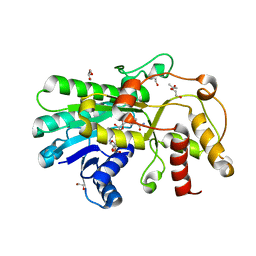 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
8I99
 
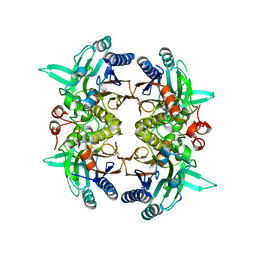 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | Descriptor: | N-carbamoyl-D-amino-acid hydrolase | | Authors: | Hu, J.M, Ni, Y, Xu, G.C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
6LCG
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Liu, Y.F, Ni, Y, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
6LEI
 
 | |
6LED
 
 | |
6LE2
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
