8HIL
 
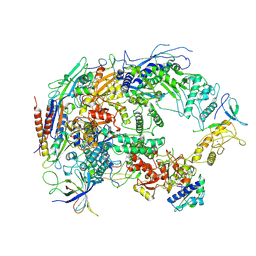 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8HIM
 
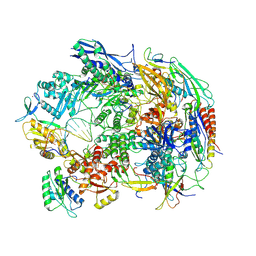 | | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | | Descriptor: | DNA (34-MER), DNA-directed RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, ... | | Authors: | Hu, H, Xie, G, Du, X, Du, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
1BPS
 
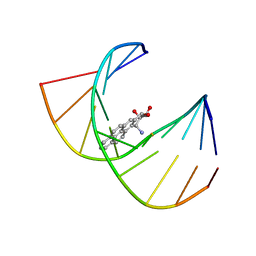 | | MINOR CONFORMER OF A BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*(BAP)AP*CP*GP*AP*G)-3') | | Authors: | Schwartz, J.S, Rice, J.S, Luxon, B.A, Sayer, J.M, Xie, G, Yeh, H.J.C, Liu, X, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the minor conformer of a DNA duplex containing a dG mismatch opposite a benzo[a]pyrene diol epoxide/dA adduct: glycosidic rotation from syn to anti at the modified deoxyadenosine.
Biochemistry, 36, 1997
|
|
1VE7
 
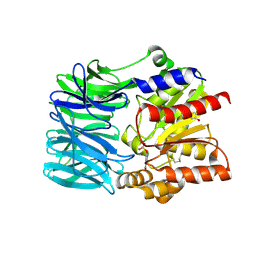 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
1VE6
 
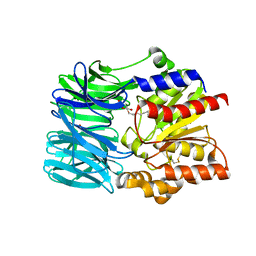 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
5JDB
 
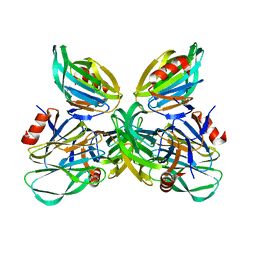 | | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens | | Descriptor: | Outer capsid protein VP4 | | Authors: | Sun, X, Guo, N, Li, D, Zhou, Y, Jin, M, Xie, G, Pang, L, Zhang, Q, Cao, Y, Duan, Z. | | Deposit date: | 2016-04-16 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens.
Virology, 495, 2016
|
|
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
2J4Y
 
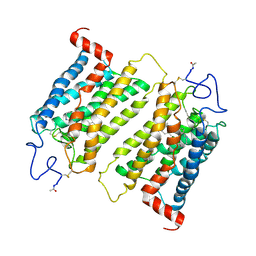 | | Crystal structure of a rhodopsin stabilizing mutant expressed in mammalian cells | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, RHODOPSIN | | Authors: | Standfuss, J, Xie, G, Edwards, P.C, Burghammer, M, Oprian, D.D, Schertler, G.F.X. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of a Thermally Stable Rhodopsin Mutant.
J.Mol.Biol., 372, 2007
|
|
2X72
 
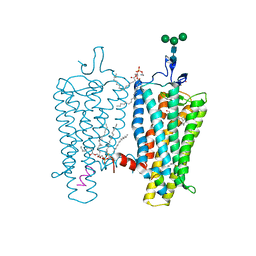 | | CRYSTAL STRUCTURE OF THE CONSTITUTIVELY ACTIVE E113Q,D2C,D282C RHODOPSIN MUTANT WITH BOUND GALPHACT PEPTIDE. | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ACETATE ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Standfuss, J, Edwards, P.C, Dantona, A, Fransen, M, Xie, G, Oprian, D.D, Schertler, G.F.X. | | Deposit date: | 2010-02-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structural Basis of Agonist Induced Activation in Constitutively Active Rhodopsin
Nature, 471, 2011
|
|
6AKJ
 
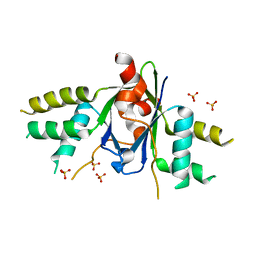 | | The crystal structure of EMC complex | | Descriptor: | Enhancer of rudimentary homolog,YTH domain-containing protein mmi1 fusion protein, SULFATE ION | | Authors: | Li, F. | | Deposit date: | 2018-09-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing.
Nat Commun, 10, 2019
|
|
9ATN
 
 | |
6LYH
 
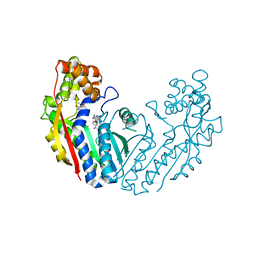 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
6LYI
 
 | |
4A4M
 
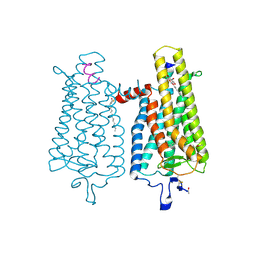 | | Crystal structure of the light-activated constitutively active N2C, M257Y,D282C rhodopsin mutant in complex with a peptide resembling the C-terminus of the Galpha-protein subunit (GaCT) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GUANINE NUCLEOTIDE-BINDING PROTEIN G(T) SUBUNIT ALPHA-3, ... | | Authors: | Deupi, X, Edwards, P, Singhal, A, Nickle, B, Oprian, D.D, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Stabilized G Protein Binding Site in the Structure of Constitutively Active Metarhodopsin-II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7N7V
 
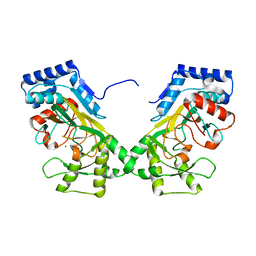 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
7YHQ
 
 | |
7YHO
 
 | |
7YHP
 
 | |
3C9M
 
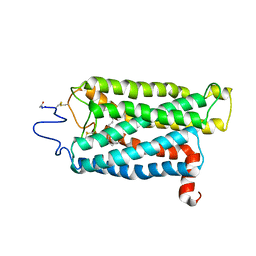 | |
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA9
 
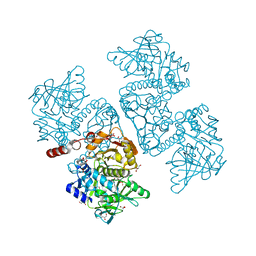 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
2HU7
 
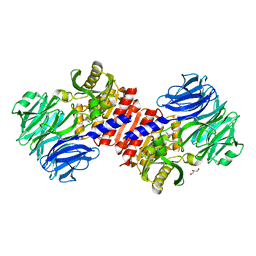 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | ACETYL GROUP, Acylamino-acid-releasing enzyme, GLYCEROL, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU8
 
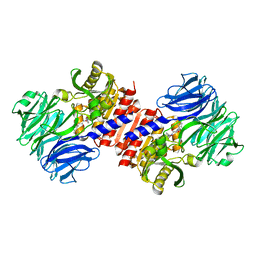 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | 2-AMINOBENZOIC ACID, Acylamino-acid-releasing enzyme, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU5
 
 | | Binding of inhibitors by Acylaminoacyl-peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
