7OML
 
 | |
7OLH
 
 | |
7OJR
 
 | |
7OJS
 
 | |
6EHC
 
 | |
6EHD
 
 | |
6EHE
 
 | |
6EHF
 
 | |
6EHB
 
 | |
5OYK
 
 | |
2NSM
 
 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
1JFQ
 
 | | ANTIGEN-BINDING FRAGMENT OF THE MURINE ANTI-PHENYLARSONATE ANTIBODY 36-71, "FAB 36-71" | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF ANTI-PHENYLARSONATE ANTIBODY | | Authors: | Parhami-Seren, B, Viswanathan, M, Strong, R.K, Margolies, M.N. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mutants of high-affinity and low-affinity
p-azophenylarsonate-specific antibodies generated by alanine
scanning of heavy chain
complementarity-determining region 2.
J.Immunol., 167, 2001
|
|
2N3D
 
 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
2XIT
 
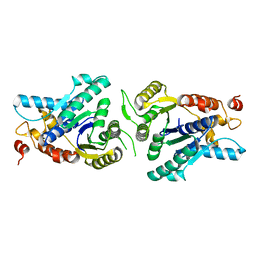 | | Crystal structure of monomeric MipZ | | Descriptor: | MIPZ | | Authors: | Kiekebusch, D, Michie, K.A, Essen, L.O, Lowe, J, Thanbichler, M. | | Deposit date: | 2010-06-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Localized Dimerization and Nucleoid Binding Drive Gradient Formation by the Bacterial Cell Division Inhibitor Mipz.
Mol.Cell, 46, 2012
|
|
5O77
 
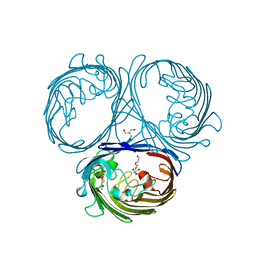 | | Klebsiella pneumoniae OmpK35 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | Authors: | van den berg, B, Pathania, M, Zahn, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5O79
 
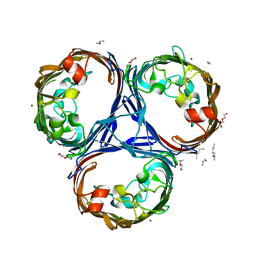 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
4ACB
 
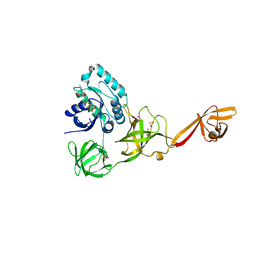 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH THE GTP ANALOGUE GPPNHP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4AC9
 
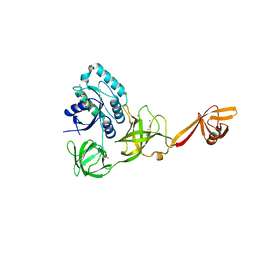 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH GDP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4ACA
 
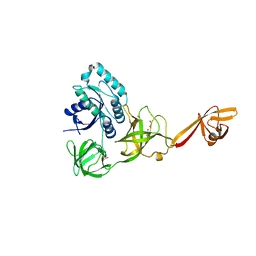 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS, APO FORM | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
5FVN
 
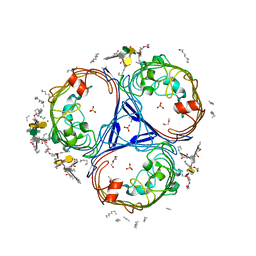 | | X-ray crystal structure of Enterobacter cloacae OmpE36 porin. | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, ... | | Authors: | Arunmanee, W, Pathania, M, Soloyova, A, Brun, A, Ridley, H, Basle, A, van den Berg, B, Lakey, J.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5T89
 
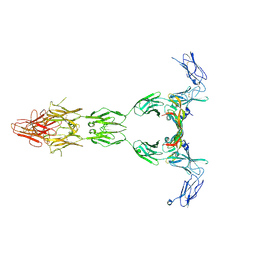 | | Crystal structure of VEGF-A in complex with VEGFR-1 domains D1-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vascular endothelial growth factor A, ... | | Authors: | Markovic-Mueller, S, Stuttfeld, E, Asthana, M, Weinert, T, Bliven, S, Goldie, K.N, Kisko, K, Capitani, G, Ballmer-Hofer, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A.
Structure, 25, 2017
|
|
4KG9
 
 | | Crystal Structure Of USP7-NTD with MCM-BP | | Descriptor: | Mini-chromosome maintenance complex-binding protein, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Luthra, N. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A role for USP7 in DNA replication.
Mol.Cell.Biol., 34, 2014
|
|
7O0N
 
 | |
4OGY
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
2NAP
 
 | | DISSIMILATORY NITRATE REDUCTASE (NAP) FROM DESULFOVIBRIO DESULFURICANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Dias, J.M, Than, M, Humm, A, Huber, R, Bourenkov, G, Bartunik, H, Bursakov, S, Calvete, J, Caldeira, J, Carneiro, C, Moura, J, Moura, I, Romao, M.J. | | Deposit date: | 1998-09-18 | | Release date: | 1999-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods.
Structure Fold.Des., 7, 1999
|
|
