6CMY
 
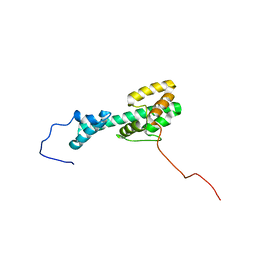 | | Solution NMR Structure Determination of Mouse Melanoregulin | | Descriptor: | Melanoregulin | | Authors: | Rout, A.K, Wu, X, Strub, M.P, Starich, M.R, Hammer III, J.A, Tjandra, N. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Melanoregulin Reveals a Role for Cholesterol Recognition in the Protein's Ability to Promote Dynein Function.
Structure, 26, 2018
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1LXE
 
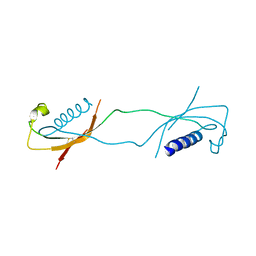 | | CRYSTAL STRUCTURE OF THE CATHELICIDIN MOTIF OF PROTEGRINS | | Descriptor: | protegrin-3 precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1N5H
 
 | | Solution structure of the cathelin-like domain of protegrins (the R87-P88 and D118-P119 amide bonds are in the cis conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
1N5P
 
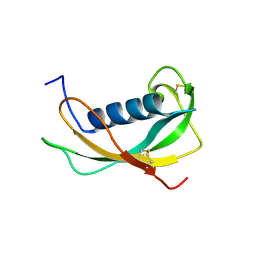 | | Solution structure of the cathelin-like domain of protegrins (all amide bonds involving proline residues are in trans conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
8EFF
 
 | | CryoEM of the soluble OPA1 tetramer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFS
 
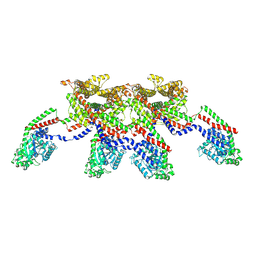 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EF7
 
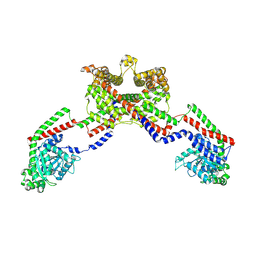 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFR
 
 | | CryoEM of the soluble OPA1 interfaces with GDP-AlFx bound from the helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EEW
 
 | | CryoEM of the soluble OPA1 dimer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFT
 
 | | CryoEM of the soluble OPA1 interfaces from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
1HP8
 
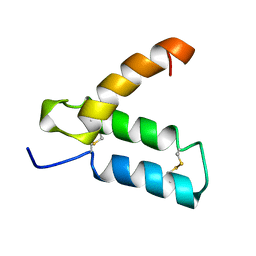 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
1G33
 
 | | CRYSTAL STRUCTURE OF RAT PARVALBUMIN WITHOUT THE N-TERMINAL DOMAIN | | Descriptor: | CALCIUM ION, PARVALBUMIN ALPHA, SULFATE ION | | Authors: | Thepaut, M, Strub, M.P, Cave, A, Baneres, J.L, Berchtold, M.W, Dumas, C, Padilla, A. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of rat parvalbumin with deleted AB domain: implications for the evolution of EF hand calcium-binding proteins and possible physiological relevance.
Proteins, 45, 2001
|
|
2HP8
 
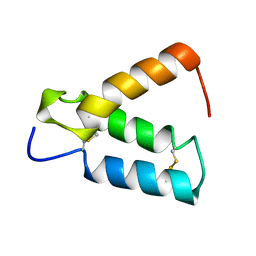 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
5VKG
 
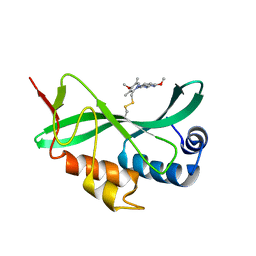 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with tenatoprazole | | Descriptor: | 4-methoxy-1-(5-methoxy-3H-imidazo[4,5-b]pyridin-2-yl)-3,5-dimethyl-2-(sulfanylmethyl)pyridin-1-ium, Tumor susceptibility gene 101 protein | | Authors: | Strickland, M, Ehrlich, L.S, Watanabe, S, Khan, M, Strub, M.-P, Luan, C.H, Powell, M.D, Leis, J, Tjandra, N, Carter, C. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Tsg101 chaperone function revealed by HIV-1 assembly inhibitors.
Nat Commun, 8, 2017
|
|
6DLU
 
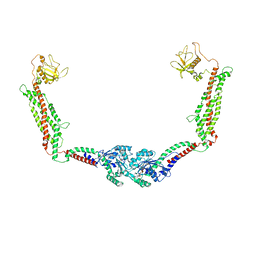 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DLV
 
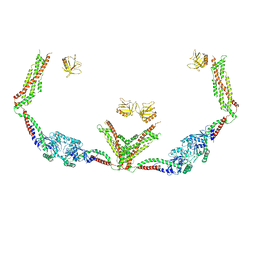 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
3F45
 
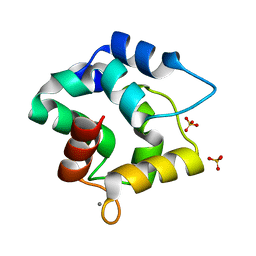 | | Structure of the R75A mutant of rat alpha-Parvalbumin | | Descriptor: | CALCIUM ION, Parvalbumin alpha, SULFATE ION | | Authors: | Hoh, F, Padilla, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removing the invariant salt bridge of parvalbumin increases flexibility in the AB-loop structure
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1AVV
 
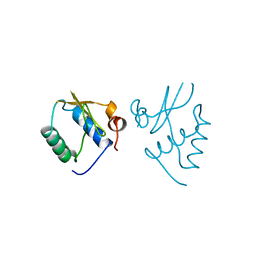 | | HIV-1 NEF PROTEIN, UNLIGANDED CORE DOMAIN | | Descriptor: | NEGATIVE FACTOR | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1997-09-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling.
Structure, 5, 1997
|
|
1AVZ
 
 | |
1AUU
 
 | |
1BU1
 
 | | SRC FAMILY KINASE HCK SH3 DOMAIN | | Descriptor: | PROTEIN (HEMOPOIETIC CELL KINASE) | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1998-09-09 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef.
Biochemistry, 37, 1998
|
|
8SZ4
 
 | |
8SZ7
 
 | |
8T0R
 
 | |
