1BG0
 
 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1M15
 
 | | Transition state structure of arginine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, MAGNESIUM ION, ... | | Authors: | Yousef, M.S, Fabiola, F, Gattis, J.L, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Refinement of the arginine kinase transition-state analogue complex at 1.2 A resolution: mechanistic insights.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LCU
 
 | | Polylysine Induces an Antiparallel Actin Dimer that Nucleates Filament Assembly: Crystal Structure at 3.5 A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Bubb, M.R, Govindasamy, L, Yarmola, E.G, Vorobiev, S.M, Almo, S.C, Somasundaram, T, Chapman, M.S, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2002-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution.
J.Biol.Chem., 277, 2002
|
|
2HZ9
 
 | | Crystal structure of Lys12Val/Asn95Val/Cys117Val mutant of human acidic fibroblast growth factor at 1.70 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-08 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spackling the Crack: Stabilizing Human Fibroblast Growth Factor-1 by Targeting the N and C terminus beta-Strand Interactions
J.Mol.Biol., 371, 2007
|
|
3M10
 
 | | Substrate-free form of Arginine Kinase | | Descriptor: | Arginine kinase, SULFATE ION | | Authors: | Yousef, M.S, Clark, S.A, Pruett, P.K, Somasundaram, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Arginine kinase: joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility.
J.Mol.Biol., 405, 2011
|
|
3JPZ
 
 | | Crystal Structure of Lombricine Kinase | | Descriptor: | Lombricine kinase, NITRATE ION | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
3JQ3
 
 | | Crystal Structure of Lombricine Kinase, complexed with substrate ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lombricine kinase | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
1RG8
 
 | |
1LP3
 
 | | The Atomic Structure of Adeno-Associated Virus (AAV-2), a Vector for Human Gene Therapy | | Descriptor: | AAV-2 capsid protein | | Authors: | Xie, Q, Bu, W, Bhatia, S, Hare, J, Somasundaram, T, Azzi, A, Chapman, M.S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2HWA
 
 | | Crystal structure of Lys12Thr/Cys117Val mutant of human acidic fibroblast growth factor at 1.65 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HW9
 
 | | Crystal structure of Lys12Cys/Cys117Val mutant of human acidic fibroblast Growth factor at 1.60 angstrom resolution. | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
2HWM
 
 | | Crystal structure of Lys12Val/Cys117Val mutant of human acidic fibroblast growth factor at 1.60 angstrom resolution | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1 | | Authors: | Dubey, V.K, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2006-08-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions.
J.Mol.Biol., 371, 2007
|
|
1IJJ
 
 | | THE X-RAY CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN RABBIT SKELETAL MUSCLE ACTIN AND LATRUNCULIN A AT 2.85 A RESOLUTION | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vorobiev, S.M, Bubb, M.R, Almo, S.C. | | Deposit date: | 2001-04-26 | | Release date: | 2002-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution
J.Biol.Chem., 277, 2002
|
|
1SD0
 
 | | Structure of arginine kinase C271A mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Gattis, J.L, Ruben, E, Fenley, M.O, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2004-02-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants
Biochemistry, 43, 2004
|
|
1P50
 
 | | Transition state structure of an Arginine Kinase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P52
 
 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
2NTD
 
 | |
1YTO
 
 | | Crystal Structure of Gly19 deletion Mutant of Human Acidic Fibroblast Growth Factor | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-02-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
1Z4S
 
 | |
1Z2V
 
 | |
2AQZ
 
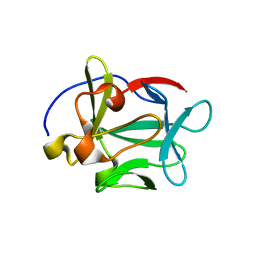 | | Crystal structure of FGF-1, S17T/N18T/G19 deletion mutant | | Descriptor: | Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
1QH4
 
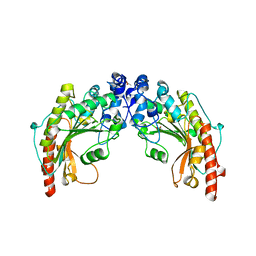 | | CRYSTAL STRUCTURE OF CHICKEN BRAIN-TYPE CREATINE KINASE AT 1.41 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CALCIUM ION, CREATINE KINASE | | Authors: | Eder, M, Schlattner, U, Becker, A, Wallimann, T, Kabsch, W, Fritz-Wolf, K. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of brain-type creatine kinase at 1.41 A resolution.
Protein Sci., 8, 1999
|
|
2ERM
 
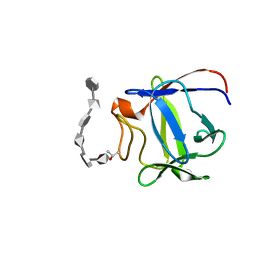 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
3CRG
 
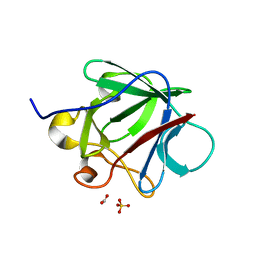 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
