3D79
 
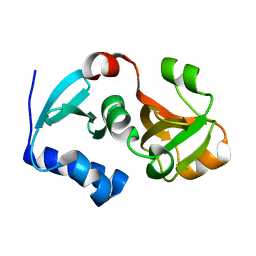 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
1VF9
 
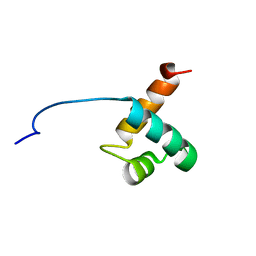 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1VFC
 
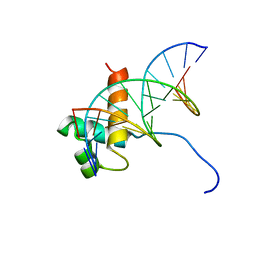 | | Solution Structure Of The DNA Complex Of Human Trf2 | | Descriptor: | Short C-rich starnd, Short G-rich strand, Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1JIB
 
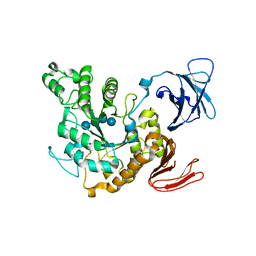 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1JL8
 
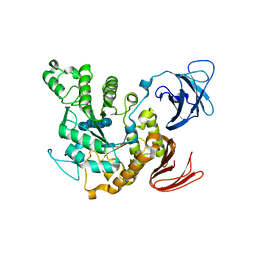 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
1JF6
 
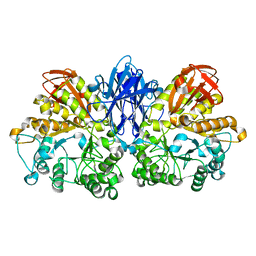 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JF5
 
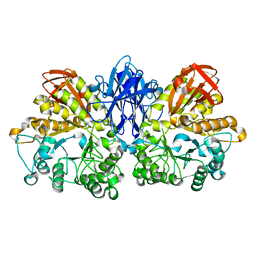 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1B7F
 
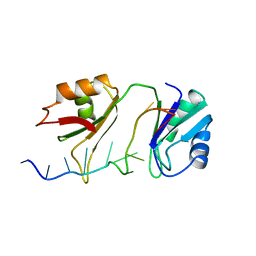 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
1BVZ
 
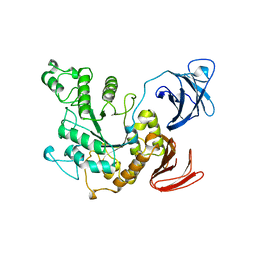 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
1WZK
 
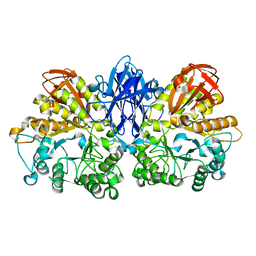 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
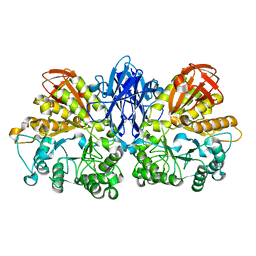 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZM
 
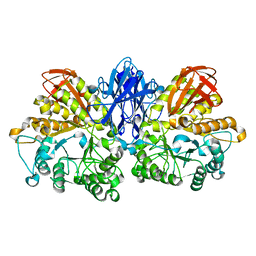 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1MBF
 
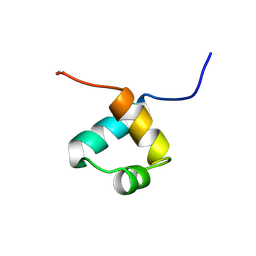 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBH
 
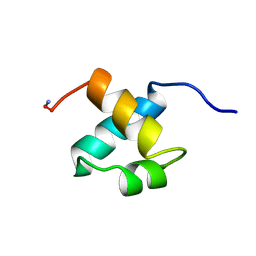 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBE
 
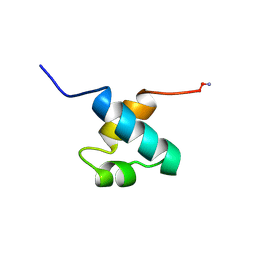 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
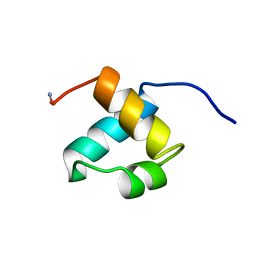 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
8I53
 
 | |
3J3Z
 
 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1MSE
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1MSF
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
