3SQG
 
 | | Crystal structure of a methyl-coenzyme M reductase purified from Black Sea mats | | Descriptor: | (17[2]S)-17[2]-methylthio-coenzyme F43, 1-THIOETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shima, S, Krueger, M, Weinert, T, Demmer, U, Thauer, R.K, Ermler, U. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically.
Nature, 481, 2011
|
|
1F07
 
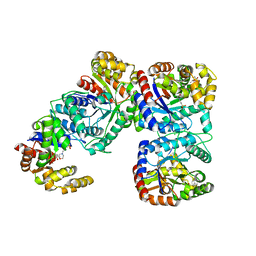 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1EZW
 
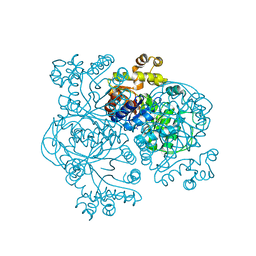 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
2KTD
 
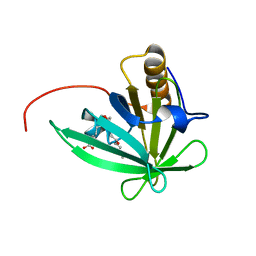 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
8JT1
 
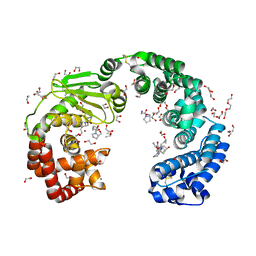 | | COLLAGENASE FROM GRIMONTIA (VIBRIO) HOLLISAE 1706B COMPLEXED WITH GLY-PRO-HYP-GLY-PRO-HYP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-mer peptide, ... | | Authors: | Ueshima, S, Yaskawa, K, Takita, T, Mikami, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the catalytic mechanism of Grimontia hollisae collagenase through structural and mutational analyses.
Febs Lett., 597, 2023
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
7CSS
 
 | |
7D37
 
 | |
2E4J
 
 | |
3DAG
 
 | | The crystal structure of [Fe]-hydrogenase holoenzyme (HMD) from METHANOCALDOCOCCUS JANNASCHII | | Descriptor: | 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Pilak, O, Warkentin, E, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2008-05-29 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site.
Science, 321, 2008
|
|
3DAF
 
 | | The crystal structure of [Fe]-hydrogenase holoenzyme (HMD) from METHANOCALDOCOCCUS JANNASCHII cocrystallized with cyanide | | Descriptor: | 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Pilak, O, Warkentin, E, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2008-05-29 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site.
Science, 321, 2008
|
|
8HR3
 
 | | [D-Cys5,Asp7,Val8,D-Lys16]-STp(5-17) | | Descriptor: | DCY-CYS-ASP-VAL-CYS-CYS-ASN-PRO-ALA-CYS-ALA-DLY-CYS | | Authors: | Shimamoto, S, Hidaka, Y, Yoshino, S, Goto, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-20 | | Method: | SOLUTION NMR | | Cite: | The Molecular Basis of Heat-Stable Enterotoxin for Vaccine Development and Cancer Cell Detection.
Molecules, 28, 2023
|
|
8HR4
 
 | |
7VSO
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with HAD16 Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2-[[(2R)-2-(3-bromanyl-5-iodanyl-phenyl)carbonyloxy-3-tetradecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, ... | | Authors: | Mizohata, E, Nakane, T, Hanashima, S. | | Deposit date: | 2021-10-27 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins.
Membranes (Basel), 11, 2021
|
|
2RQP
 
 | |
8BGR
 
 | |
6QII
 
 | | Xenon derivatization of the F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGT
 
 | | The carbon monoxide inhibition of F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGR
 
 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3F47
 
 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
1JAX
 
 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) | | Descriptor: | MAGNESIUM ION, SODIUM ION, conserved hypothetical protein | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
6S6Y
 
 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LB9
 
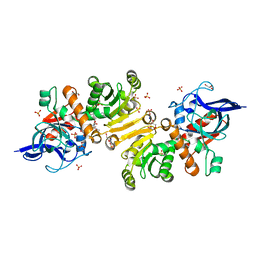 | | Structure of the T175V Etr1p mutant in the monoclinic form P21 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
8QPL
 
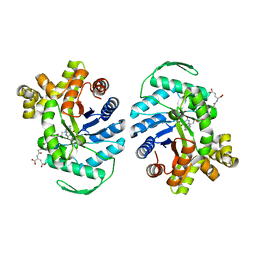 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
