3FDQ
 
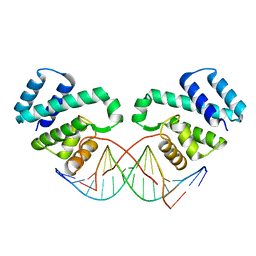 | |
3GCD
 
 | | Structure of the V. cholerae RTX cysteine protease domain in complex with an aza-Leucine peptide inhibitor | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION, ... | | Authors: | Lupardus, P.J, Garcia, K.C, Shen, A, Bogyo, M. | | Deposit date: | 2009-02-21 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin.
Nat.Chem.Biol., 5, 2009
|
|
3EEB
 
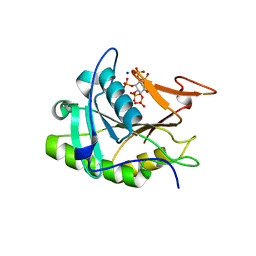 | | Structure of the V. cholerae RTX cysteine protease domain | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION | | Authors: | Lupardus, P.J, Shen, A, Bogyo, M, Garcia, K.C. | | Deposit date: | 2008-09-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
Science, 322, 2008
|
|
1C7I
 
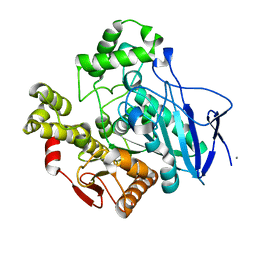 | | THERMOPHYLIC PNB ESTERASE | | Descriptor: | CALCIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE) | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7J
 
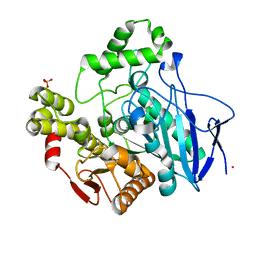 | | PNB ESTERASE 56C8 | | Descriptor: | POTASSIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE), SULFATE ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R.C. | | Deposit date: | 2000-02-21 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6NST
 
 | |
4S3G
 
 | | Structure of the F249X mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
4RV3
 
 | | Crystal structure of a pentafluoro-Phe incorporated Phosphatidylinositol-specific phospholipase C (H258X)from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2014-11-24 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
1QE3
 
 | | PNB ESTERASE | | Descriptor: | PARA-NITROBENZYL ESTERASE, SULFATE ION, ZINC ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | Deposit date: | 1999-07-12 | | Release date: | 1999-07-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4F2U
 
 | | Structure of the N254Y/H258Y double mutant of the Phosphatidylinositol-Specific Phospholipase C from S.aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4F2T
 
 | | Modulation of S.aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4F2B
 
 | | Modulation of S.Aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4I8Y
 
 | | Structure of the unliganded N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9J
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I90
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to choline | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9M
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9T
 
 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
3PEE
 
 | |
4LWG
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWH
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LWE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4L94
 
 | | Crystal structure of Human Hsp90 with S46 | | Descriptor: | (4-hydroxyphenyl)(4-methylpiperazin-1-yl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LWI
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWF
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ3 | | Descriptor: | 4-(5-amino-1,2-oxazol-3-yl)-6-(propan-2-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
