7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7ASB
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7OU5
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite Dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OWI
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU9
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU7
 
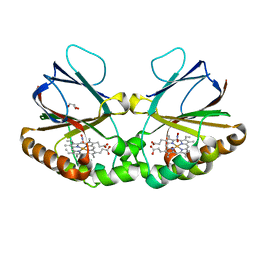 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUA
 
 | | Crystal structure of dimeric chlorite dismutase variant R127K (CCld R127K) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUY
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
2GED
 
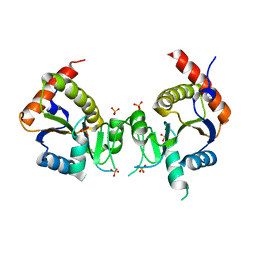 | |
7TXD
 
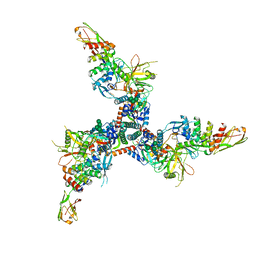 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6F26
 
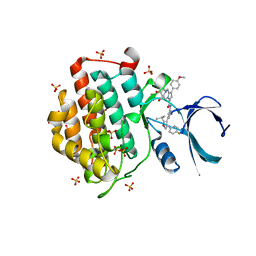 | | Crystal structure of human Casein Kinase I delta in complex with compound 31b | | Descriptor: | (9~{S},10~{S},11~{R})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
6F1W
 
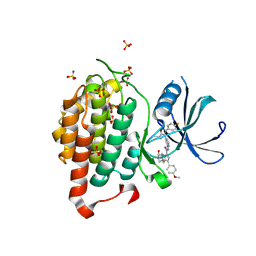 | | Crystal structure of human Casein Kinase I delta in complex with compound 31a | | Descriptor: | (9~{R},10~{R},11~{S})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
7RA5
 
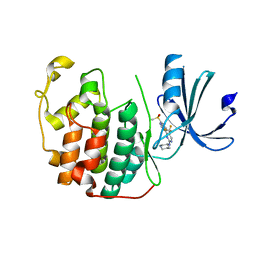 | | CDK2 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-[7-(methanesulfonyl)-1H-indol-3-yl]-N-[(3S)-piperidin-3-yl]-5-(trifluoromethyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Marineau, J.J, Malojcic, G. | | Deposit date: | 2021-06-30 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of SY-5609: A Selective, Noncovalent Inhibitor of CDK7.
J.Med.Chem., 65, 2022
|
|
7Z7C
 
 | |
6RR5
 
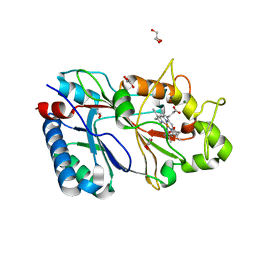 | | Structure of 50% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR1
 
 | | Structure of 10% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR8
 
 | | Structure of 100% reduced KpDyP (final wedges) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR4
 
 | | Structure of 25% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RR6
 
 | | Structure of 100% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RQY
 
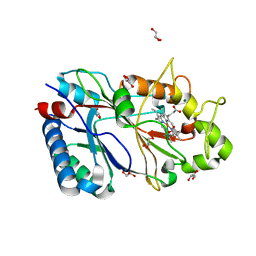 | | Structure of % reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
8AED
 
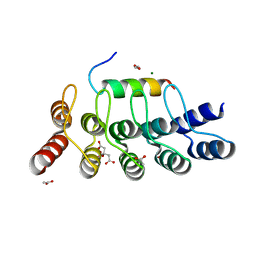 | |
6RPE
 
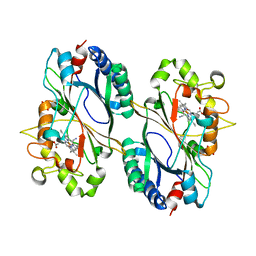 | | Structure of 5% reduced KpDyP in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, Iron-dependent peroxidase, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RPD
 
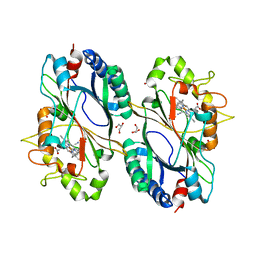 | | Structure of ferrous KpDyP in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, Iron-dependent peroxidase, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
5A10
 
 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group C2) | | Descriptor: | ACETATE ION, SODIUM ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
5A11
 
 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group P21) | | Descriptor: | IODIDE ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
