5KTZ
 
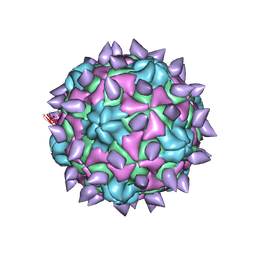 | | expanded poliovirus in complex with VHH 12B | | Descriptor: | Genome polyprotein, VHH 12B, VP1, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KU0
 
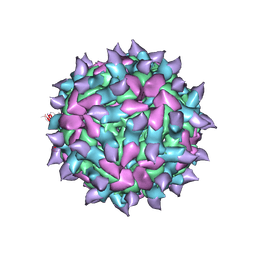 | | expanded poliovirus in complex with VHH 17B | | Descriptor: | VHH 17B, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KU2
 
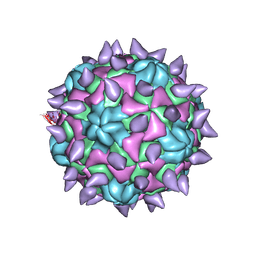 | | expanded poliovirus in complex with VHH 7A | | Descriptor: | VHH 7A, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
5KWL
 
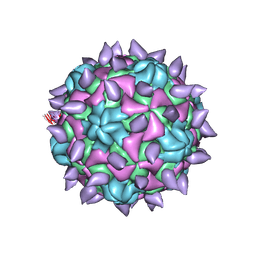 | | expanded poliovirus in complex with VHH 10E | | Descriptor: | VHH 10E, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-18 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
3J8F
 
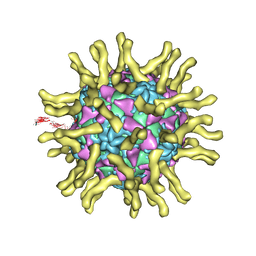 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3JBF
 
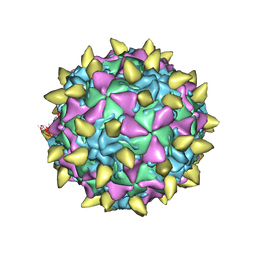 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBE
 
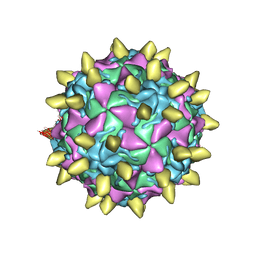 | | Complex of poliovirus with VHH PVSS8A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBD
 
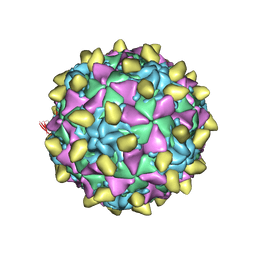 | | Complex of poliovirus with VHH PVSP6A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBC
 
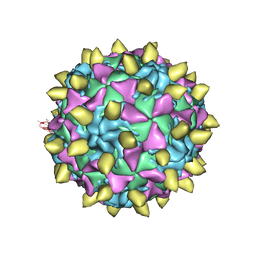 | | Complex of Poliovirus with VHH PVSP29F | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBG
 
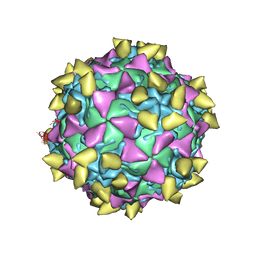 | | Complex of poliovirus with VHH PVSS21E | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3J9F
 
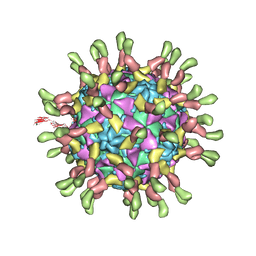 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
4PC8
 
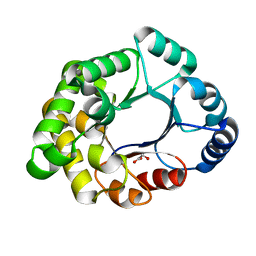 | |
4PCF
 
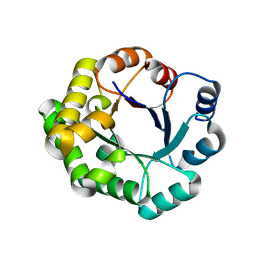 | |
2MI6
 
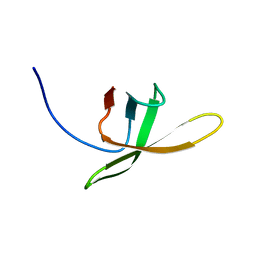 | |
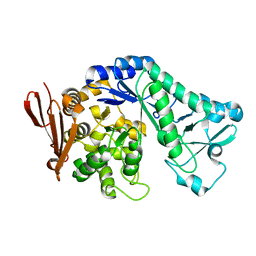
5MAN
 
 | |
5M9X
 
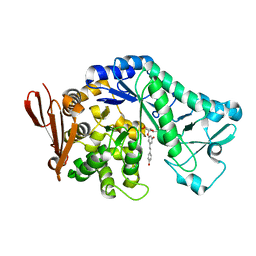 | | Structure of sucrose phosphorylase from Bifidobacterium adolescentis bound to glycosylated resveratrol | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[3-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-5-oxidanyl-phenoxy]oxane-3,4 ,5-triol, Sucrose phosphorylase | | Authors: | Grimm, C, Kraus, M. | | Deposit date: | 2016-11-02 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Switching enzyme specificity from phosphate to resveratrol glucosylation.
Chem. Commun. (Camb.), 53, 2017
|
|
8JFC
 
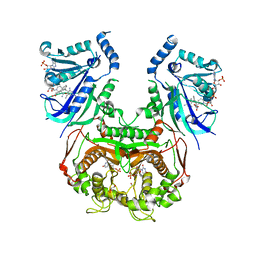 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 6 (B21591), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[3-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
8JFB
 
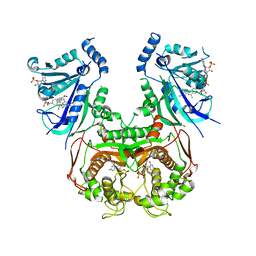 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 4 (B21588), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-[4-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
8JFD
 
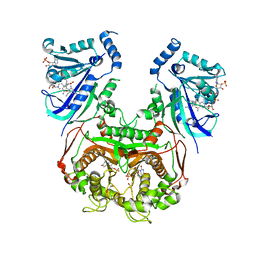 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 8 (B21594), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-[3-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
5C8B
 
 | |
7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Z
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
