1R6R
 
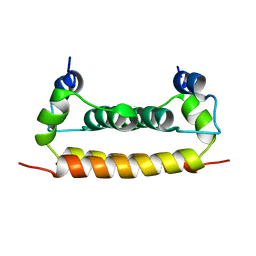 | | Solution Structure of Dengue Virus Capsid Protein Reveals a New Fold | | Descriptor: | Genome polyprotein | | Authors: | Ma, L, Jones, C.T, Groesch, T.D, Kuhn, R.J, Post, C.B. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dengue virus capsid protein reveals another fold
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2BTB
 
 | |
2BTA
 
 | |
3BTB
 
 | |
2FCI
 
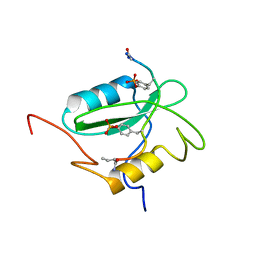 | | Structural basis for the requirement of two phosphotyrosines in signaling mediated by Syk tyrosine kinase | | Descriptor: | C-termainl SH2 domain from phospholipase C-gamma-1 comprising residues 663-759, Doubly phosphorylated peptide derived from Syk kinase comprising residues 338-350 | | Authors: | Groesch, T.D, Zhou, F, Mattila, S, Geahlen, R.L, Post, C.B. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by syk tyrosine kinase
J.Mol.Biol., 356, 2006
|
|
1EOQ
 
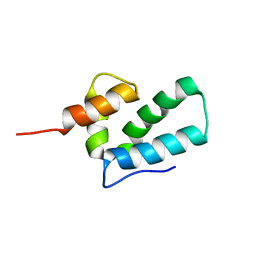 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1EM9
 
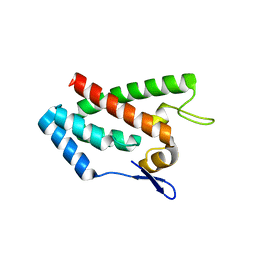 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: N-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27, MAGNESIUM ION | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1CV9
 
 | | NMR STUDY OF ITAM PEPTIDE SUBSTRATE | | Descriptor: | IG-ALPHA ITAM PEPTIDE | | Authors: | Gaul, B.S, Harrison, M.L, Geahlen, R.L, Post, C.B. | | Deposit date: | 1999-08-23 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Substrate recognition by the Lyn protein-tyrosine kinase. NMR structure of the immunoreceptor tyrosine-based activation motif signaling region of the B cell antigen receptor.
J.Biol.Chem., 275, 2000
|
|
2LCT
 
 | |
2MC1
 
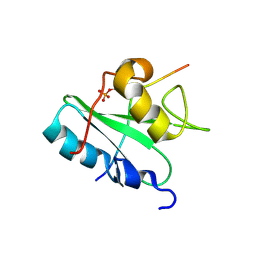 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
1JDY
 
 | |
1C47
 
 | |
1C4G
 
 | |
1LXT
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE PHOSPHOGLUCOMUTASE FROM RABBIT | | Descriptor: | CADMIUM ION, PHOSPHOGLUCOMUTASE (DEPHOSPHO FORM), SULFATE ION | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
3PMG
 
 | |
1VKL
 
 | | RABBIT MUSCLE PHOSPHOGLUCOMUTASE | | Descriptor: | NICKEL (II) ION, PHOSPHOGLUCOMUTASE | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural changes at the metal ion binding site during the phosphoglucomutase reaction.
Biochemistry, 32, 1993
|
|
