2QJR
 
 | | dipepdyl peptidase IV in complex with inhibitor PZF | | Descriptor: | (3R,4S)-1-[6-(6-METHOXYPYRIDIN-3-YL)PYRIMIDIN-4-YL]-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shenping, L. | | Deposit date: | 2007-07-09 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | (3R,4S)-4-(2,4,5-Trifluorophenyl)-pyrrolidin-3-ylamine inhibitors of dipeptidyl peptidase IV: synthesis, in vitro, in vivo, and X-ray crystallographic characterization.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4QR8
 
 | | Crystal Structure of E coli pepQ | | Descriptor: | MAGNESIUM ION, Xaa-Pro dipeptidase | | Authors: | Pingwei, L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of substrate selectivity of E. coli prolidase.
Plos One, 9, 2014
|
|
4JWN
 
 | | Ternary complex of D256A mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
4JWM
 
 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
6CFE
 
 | |
6CBU
 
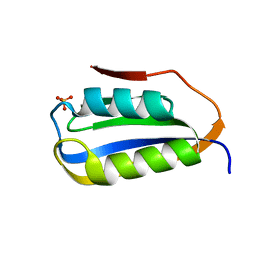 | | Crystal structure of C4S3: A computationally designed immunogen to target Carbohydrate-Occluded Epitopes on the HIV envelope | | Descriptor: | Acylphosphatase-1, SULFATE ION | | Authors: | Zhu, C, Ke, H.M, Swanstrom, R, Dokholyan, N.V. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed carbohydrate-occluded epitopes elicit HIV-1 Env-specific antibodies.
Nat Commun, 10, 2019
|
|
7PGE
 
 | | copper transporter PcoB | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Copper resistance protein B, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, P, Gourdon, P.E. | | Deposit date: | 2021-08-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PcoB is a defense outer membrane protein that facilitates cellular uptake of copper.
Protein Sci., 31, 2022
|
|
5M1U
 
 | |
1AWY
 
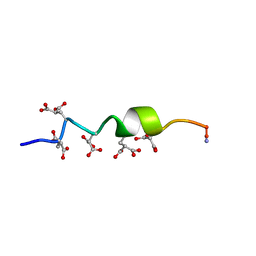 | | NMR STRUCTURE OF CALCIUM BOUND CONFORMER OF CONANTOKIN G, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CONANTOXIN G | | Authors: | Rigby, A.C, Baleja, J.D, Leping, L, Pedersen, L.G, Furie, B.C, Furie, B. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus.
Biochemistry, 36, 1997
|
|
6X59
 
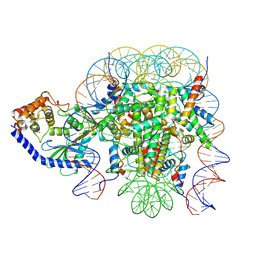 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6X5A
 
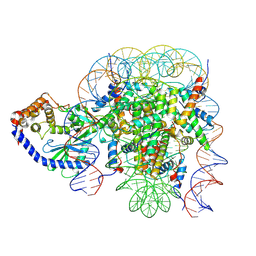 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
7VS4
 
 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
