7ZCK
 
 | | Room temperature crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Phosphonate ABC type transporter/ substrate binding component | | Authors: | Mikolajek, H, Shah, B.S, Paulsen, I.T, Sandy, J, Sanchez-Weatherby, J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
7UG8
 
 | | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium | | Descriptor: | 1,2-ETHANEDIOL, 2-oxopentanoic acid, CALCIUM ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium
To Be Published
|
|
7S6F
 
 | | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative urea ABC transporter, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium
To Be Published
|
|
7S6E
 
 | | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Mykhaylyk, V, Orr, C.M, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium
To Be Published
|
|
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
6WPM
 
 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
6WPN
 
 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Substrate-binding protein | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
4J2O
 
 | | Crystal structure of NADP-bound WbjB from A. baumannii community strain D1279779 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-N-acetylglucosamine 4,6-dehydratase/5-epimerase | | Authors: | Shah, B.S, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Crystal structure of a UDP-GlcNAc epimerase for surface polysaccharide biosynthesis in Acinetobacter baumannii.
Plos One, 13, 2018
|
|
4IUY
 
 | | Crystal structure of short-chain dehydrogenase/reductase (apo-form) from A. baumannii clinical strain WM99C | | Descriptor: | Short chain dehydrogenase/reductase | | Authors: | Shah, B.S, Tetu, S.G, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure of a short-chain dehydrogenase/reductase (SDR) within a genomic island from a clinical strain of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8A9D
 
 | | Multicrystal room temperature structure of Lysozyme collected using a double multilayer monochromator. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Sandy, J, Cheruvara, H, Mikolajek, H, Sanchez-Weatherby, J. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
8AR9
 
 | |
8AR6
 
 | |
8BRK
 
 | |
8BRL
 
 | |
8CIF
 
 | | Bovine naive ultralong antibody AbD08 collected at 293K | | Descriptor: | Heavy chain, Light chain | | Authors: | Clarke, J.D, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
3GK6
 
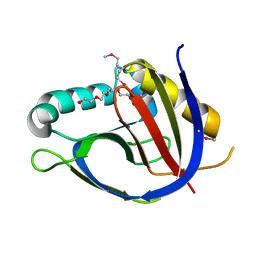 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
6RVO
 
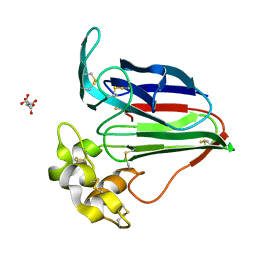 | |
6SEL
 
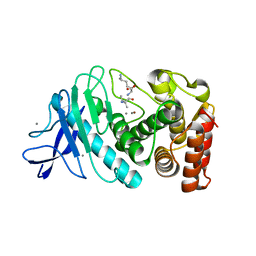 | |
6SVA
 
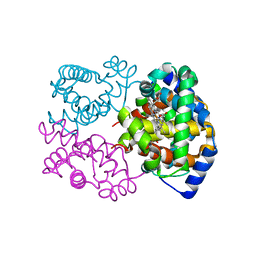 | | Multicrystal structure of equine Haemoglobin at room temperature using a multilayer monochromator. | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sandy, J, Sanchez-Weatherby, J, Mikolajek, H, Lewis, G, Angus, R. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature crystallography at VMXi
Iucrj, 2023
|
|
6RZP
 
 | | Multicrystal structure of Proteinase K at room temperature using a multilayer monochromator. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sandy, J, Sandy, E, Sanchez-Weatherby, J, Mikolajek, H. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature crystallography at VMXi
Iucrj, 2023
|
|
