8X6M
 
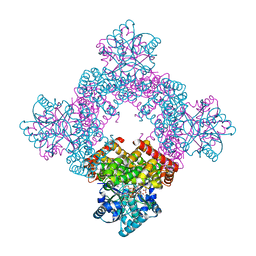 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
8GOB
 
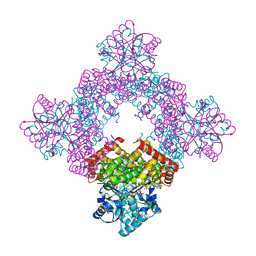 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
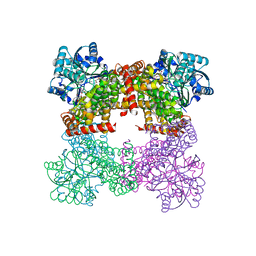 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
5MHE
 
 | |
5MHG
 
 | |
2RG0
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotetraose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RFY
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellobiose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RFW
 
 | | Crystal Structure of Cellobiohydrolase from Melanocarpus albomyces | | Descriptor: | Cellulose 1,4-beta-cellobiosidase | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2RFZ
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotriose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
2C1P
 
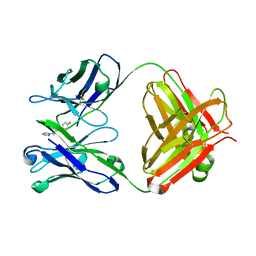 | | Fab-fragment of enantioselective antibody complexed with finrozole | | Descriptor: | 4-[(1S,2R)-3-(4-FLUOROPHENYL)-2-HYDROXY-1-(1H-1,2,4-TRIAZOL-1-YL)PROPYL]BENZONITRILE, IGH-4 PROTEIN, IGK-C PROTEIN | | Authors: | Parkkinen, T, Nevanen, T.K, Koivula, A, Rouvinen, J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of an Enantioselective Fab-Fragment in Free and Complex Forms.
J.Mol.Biol., 357, 2006
|
|
2C1O
 
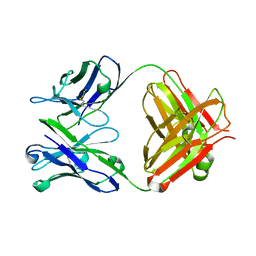 | | ENAIIHis Fab fragment in the free form | | Descriptor: | IGH-4 PROTEIN, IGK-C PROTEIN | | Authors: | Parkkinen, T, Nevanen, T.K, Koivula, A, Rouvinen, J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of an Enantioselective Fab-Fragment in Free and Complex Forms.
J.Mol.Biol., 357, 2006
|
|
3RFV
 
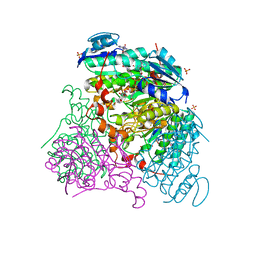 | | Crystal structure of Uronate dehydrogenase from Agrobacterium tumefaciens complexed with NADH and product | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-galactaro-1,5-lactone, PHOSPHATE ION, ... | | Authors: | Parkkinen, T, Rouvinen, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Uronate Dehydrogenase from Agrobacterium tumefaciens.
J.Biol.Chem., 286, 2011
|
|
3RFX
 
 | |
3RFT
 
 | |
6LE5
 
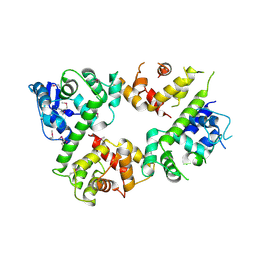 | | Crystal structure of the mitochondrial calcium uptake 1 and 2 heterodimer (MICU1-MICU2 heterodimer) in an apo state | | Descriptor: | Calcium uptake protein 1, mitochondrial, Calcium uptake protein 2 | | Authors: | Park, J, Lee, Y, Park, T, Kang, J.Y, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift.
Iucrj, 7, 2020
|
|
7YGW
 
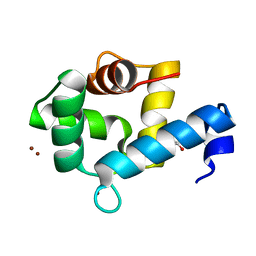 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGV
 
 | | Crystal structure of the Ca2+-bound EFhd1/Swiprosin-2 | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Ynag, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGY
 
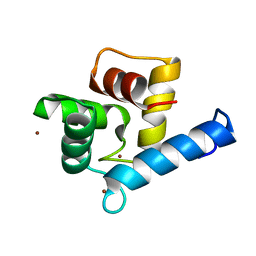 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
2DUV
 
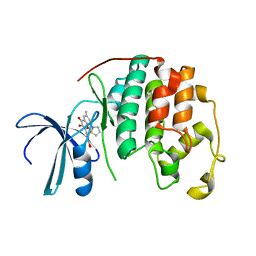 | | Structure of CDK2 with a 3-hydroxychromones | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)-8-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-3-HYDROXY-6-METHYL-4H-CHROMEN-4-ONE, Cell division protein kinase 2 | | Authors: | Kim, K.H, Lee, J, Park, T, Jeong, S, Hong, C. | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Hydroxychromones as cyclin-dependent kinase inhibitors: synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7CLT
 
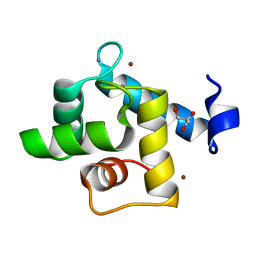 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
4UR7
 
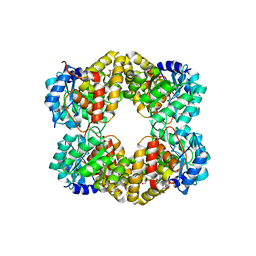 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
8RTW
 
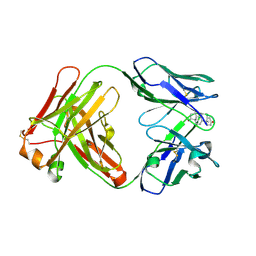 | | Crystal Structure of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | Anti-testosterone Fab 220 heavy chain, Anti-testosterone Fab 220 light chain, TESTOSTERONE | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 2024
|
|
8RTX
 
 | |
5A04
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
