6CW2
 
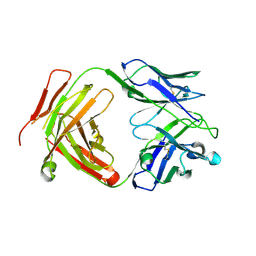 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CW3
 
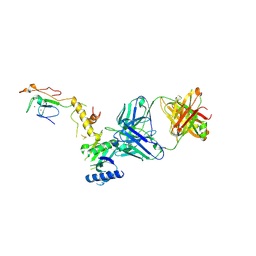 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4XTR
 
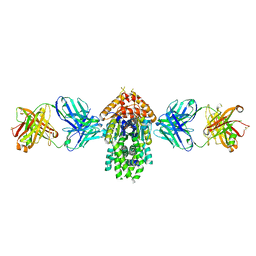 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
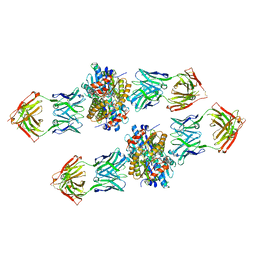 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XWO
 
 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XMN
 
 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
5CWV
 
 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWS
 
 | | Crystal structure of the intact Chaetomium thermophilum Nsp1-Nup49-Nup57 channel nucleoporin heterotrimer bound to its Nic96 nuclear pore complex attachment site | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP49, ... | | Authors: | Bley, C.J, Petrovic, S, Paduch, M, Lu, V, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWW
 
 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
6XMI
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI2
 
 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI1
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3PGF
 
 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8JNK
 
 | |
8JNR
 
 | |
4G7V
 
 | |
4G7Y
 
 | |
4G80
 
 | |
5CWT
 
 | |
4JQ5
 
 | |
4JNU
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
