4QC9
 
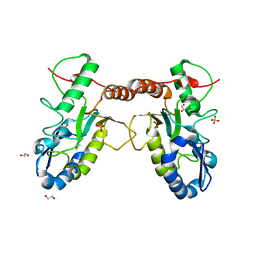 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
4QCA
 
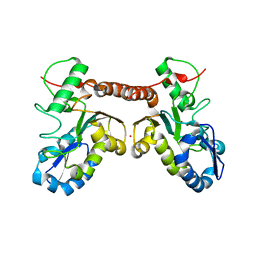 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant R167AD4 | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of three recombinant mutants of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
4IRB
 
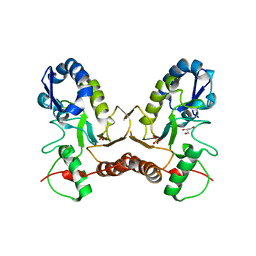 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
2RQR
 
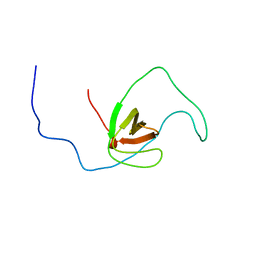 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | Descriptor: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | Authors: | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2ROG
 
 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
3A98
 
 | | Crystal structure of the complex of the interacting regions of DOCK2 and ELMO1 | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Sekine, S, Ito, T, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B13
 
 | | Crystal structure of the DHR-2 domain of DOCK2 in complex with Rac1 (T17N mutant) | | Descriptor: | Dedicator of cytokinesis protein 2, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-06-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VHL
 
 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
