3VGW
 
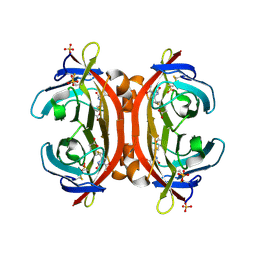 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VHH
 
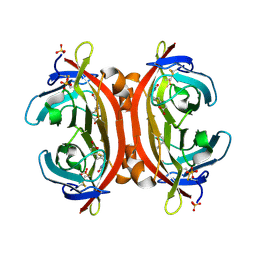 | | Crystal structure of DiMe-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1,3-dimethyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VHI
 
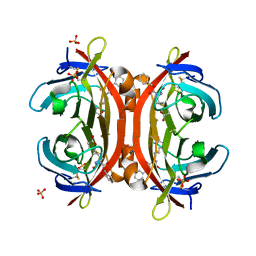 | | Crystal structure of monoZ-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{(3aS,4S,6aR)-1-[(benzyloxy)carbonyl]-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl}pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VHM
 
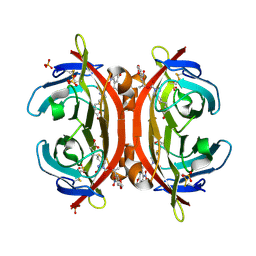 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
2JX1
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DCP*DGP*DAP*DAP*DAP*DGP*DTP*DTP*DCP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DGP*DAP*DAP*DCP*DTP*DTP*DTP*DCP*DGP*DGP*DT)-3'), Myelin transcription factor 1 | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA
To be Published
|
|
1V50
 
 | | Solution structure of phosphorylated N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
1V4Z
 
 | | Solution structure of the N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
6XVU
 
 | |
6KX8
 
 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX7
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-1 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX5
 
 | |
6KX6
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with KL101 compound | | Descriptor: | Cryptochrome-1, ~{N}-[2-(2,4-dimethylphenyl)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]-3,4-dimethyl-benzamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX4
 
 | |
5N9Q
 
 | | Structure of A. thaliana RCD1(468-567) | | Descriptor: | Inactive poly [ADP-ribose] polymerase RCD1 | | Authors: | Tossavainen, H, Hellman, M, Vainonen, J, Kangasjarvi, J, Permi, P. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors.
Elife, 8, 2019
|
|
