6U42
 
 | | Natively decorated ciliary doublet microtubule | | Descriptor: | DC1, DC2, DC3, ... | | Authors: | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4Z5Q
 
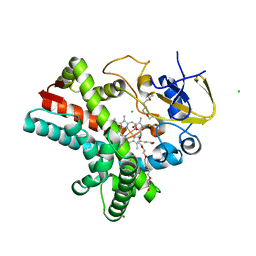 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
3OFT
 
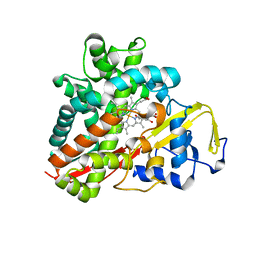 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3OFU
 
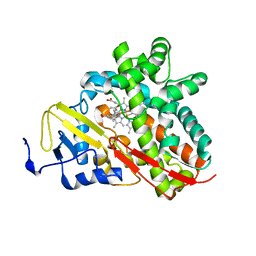 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
7CU6
 
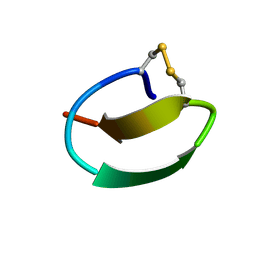 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
5BP8
 
 | | ent-Copalyl diphosphate synthase from Streptomyces platensis | | Descriptor: | 1,2-ETHANEDIOL, Ent-copalyl diphosphate synthase, SULFATE ION | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Ma, M, Chang, C.-Y, Shen, B, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
6LK3
 
 | | The Functional Characterization and Crystal Structure of Type II Peptidyl Carrier Protein ColA1a in Collismycins Biosynthesis | | Descriptor: | Putative free-standing acyl carrier protein | | Authors: | Ma, X.Y, Wang, G.Y, Liu, T, Chi, C.B, Zhang, Z.Y, Yang, D.H, Liu, W, Ma, M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional characterization and crystal structure of type II peptidyl carrier protein ColA1a in collismycins biosynthesis.
Chin.J.Chem., 38, 2020
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6M19
 
 | | Template lasso peptide C24 mutant W14F | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-02-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
7BZA
 
 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8YL9
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8YLA
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTK
 
 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7CRN
 
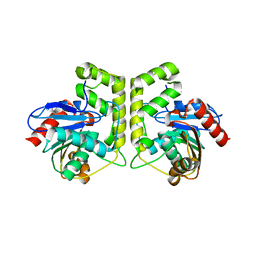 | |
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-17 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OQJ
 
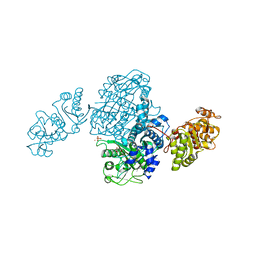 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7DXO
 
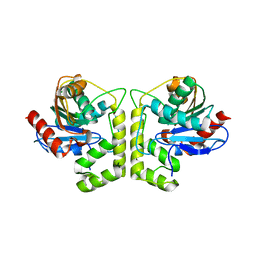 | |
