6LJJ
 
 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
7BPD
 
 | |
5XMM
 
 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
5XMF
 
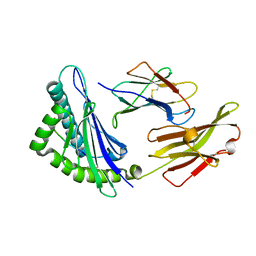 | | Crystal structure of feline MHC class I for 2,1 angstrom | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
7VWV
 
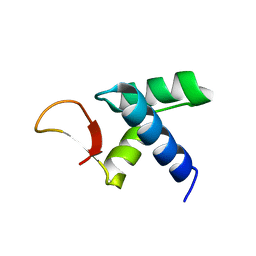 | |
7WLH
 
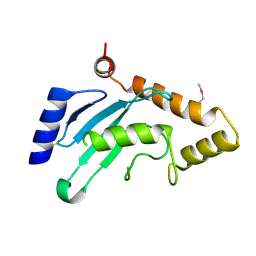 | |
6IYY
 
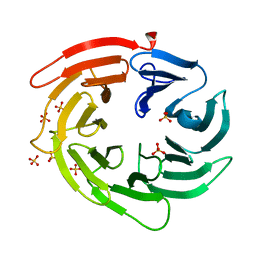 | |
6LHH
 
 | | Crystal structure of chicken 8mer-BF2*1501 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHF
 
 | | Crystal structure of chicken cCD8aa/pBF2*15:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, MHC class I, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHG
 
 | | Crystal structure of chicken cCD8aa/pBF2*04:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, IE8 peptide, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
8XR3
 
 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR4
 
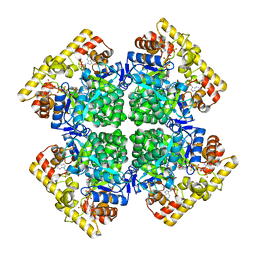 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8T60
 
 | |
6KWO
 
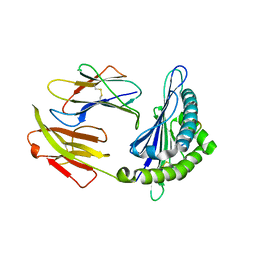 | | Crystal structure of pSLA-1*1301 complex with mutant epitope ESDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6KWL
 
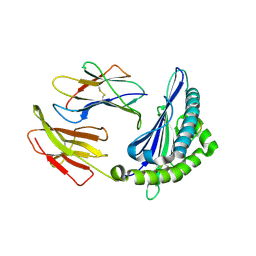 | | Crystal structure of pSLA-1*0401(R156A) complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6KWK
 
 | | Crystal structure of pSLA-1*0401 complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6LIR
 
 | | crystal structure of chicken TCR for 2.0 | | Descriptor: | TCR alpha chain, TCR beta chain | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural and Biophysical Insights into the TCR alpha beta Complex in Chickens.
Iscience, 23, 2020
|
|
6LNL
 
 | | ASFV core shell protein p15 | | Descriptor: | 60 kDa polyprotein | | Authors: | Guo, F, Shi, Y, Peng, G. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9286 Å) | | Cite: | The structural basis of African swine fever virus core shell protein p15 binding to DNA.
Faseb J., 35, 2021
|
|
6U38
 
 | | PCSK9 in complex with a Fab and compound 8 | | Descriptor: | 2-fluoro-4-{[(1R)-1-methyl-6-{[(2S)-oxan-2-yl]methoxy}-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6U2N
 
 | | PCSK9 in complex with compound 4 | | Descriptor: | 4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6UKM
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound MSA-2 | | Descriptor: | 4-(5,6-dimethoxy-1-benzothiophen-2-yl)-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-05 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
