6CRS
 
 | |
5UK1
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
2I8N
 
 | |
5JZG
 
 | | CryoEM structure of the native empty particle of a human rhinovirus C | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Liu, Y, Hill, M.G, Klose, T, Chen, Z, Watters, K.E, Jiang, W, Palmenberg, A.C, Rossmann, M.G. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QQ6
 
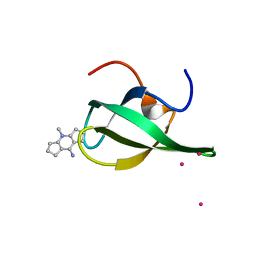 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4QQD
 
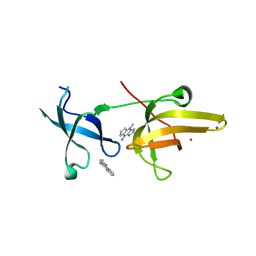 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
4RZH
 
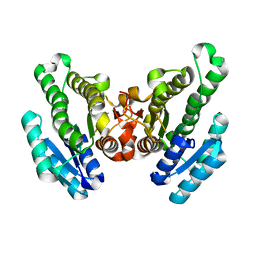 | | Crystal structure of FabG from Synechocystis sp. PCC 6803 | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Liu, Y, Xue, S. | | Deposit date: | 2014-12-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
4QQ4
 
 | | CW-type zinc finger of MORC3 in complex with the amino terminus of histone H3 | | Descriptor: | CHLORIDE ION, Histone H3.3, MORC family CW-type zinc finger protein 3, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
4ROJ
 
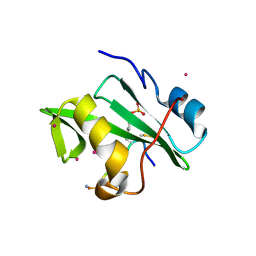 | | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide | | Descriptor: | Guanine nucleotide exchange factor VAV2, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide
TO BE PUBLISHED
|
|
1NEZ
 
 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
4RR5
 
 | | The crystal structure of Synechocystis sp. PCC 6803 Malonyl-CoA: ACP Transacylase | | Descriptor: | ACETATE ION, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Liu, Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Structural and biochemical characterization of MCAT from photosynthetic microorganism Synechocystis sp. PCC 6803 reveal its stepwise catalytic mechanism
Biochem.Biophys.Res.Commun., 457, 2015
|
|
8AHL
 
 | |
8AFH
 
 | |
8AJB
 
 | |
6X9H
 
 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
8AIA
 
 | |
8AIX
 
 | |
8AFE
 
 | |
8AFL
 
 | |
8AFM
 
 | |
3GYP
 
 | | Rtt106p | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
3GYO
 
 | | Se-Met Rtt106p | | Descriptor: | Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
4ROF
 
 | | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of WW3 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
1JS0
 
 | | Crystal Structure of 3D Domain-swapped RNase A Minor Trimer | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Gotte, G, Libonati, M, Eisenberg, D. | | Deposit date: | 2001-08-15 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the two 3D domain-swapped RNase A trimers.
Protein Sci., 11, 2002
|
|
5UHT
 
 | | Structure of the Thermotoga maritima HK853-BeF3-RR468 complex at pH 5.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, Y, Rose, J, Jiang, L, Zhou, P. | | Deposit date: | 2017-01-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
