8UDS
 
 | |
7UUR
 
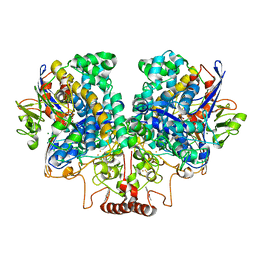 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
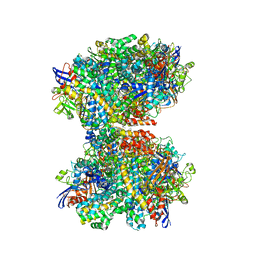 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
8DQV
 
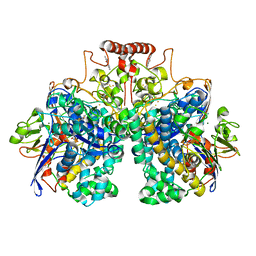 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
8OKH
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
6V4V
 
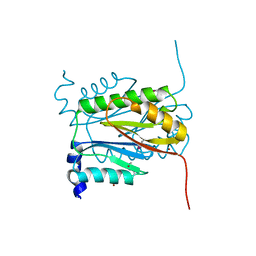 | |
5VE6
 
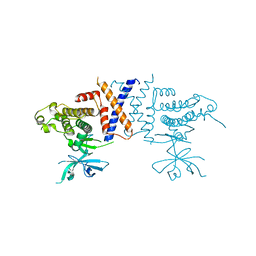 | | Crystal structure of Sugen kinase 223 | | Descriptor: | Tyrosine-protein kinase SgK223 | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association.
Nat Commun, 8, 2017
|
|
8UEM
 
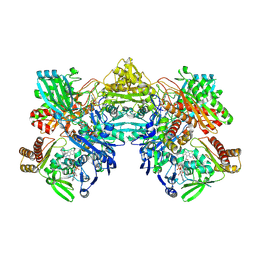 | |
5JZJ
 
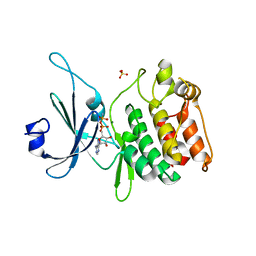 | | Crystal structure of DCLK1-KD in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
5JZN
 
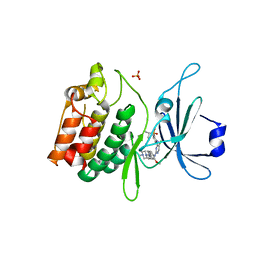 | | Crystal structure of DCLK1-KD in complex with NVP-TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, SULFATE ION, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
7KXW
 
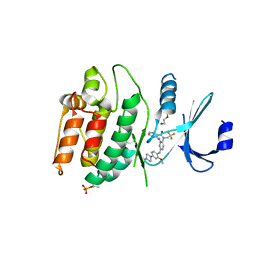 | | Crystal structure of DCLK1-KD in complex with DCLK1-IN-1 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5-(2,2,2-trifluoroethyl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase DCLK1, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
7KX8
 
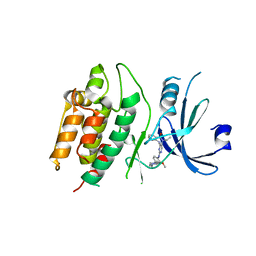 | | Crystal structure of DCLK1-Cter in complex with FMF-03-055-1 | | Descriptor: | 5-ethyl-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-03 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
7KX6
 
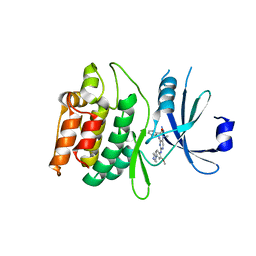 | | Crystal structure of DCLK1-KD in complex with XMD8-85 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-03 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
8DGN
 
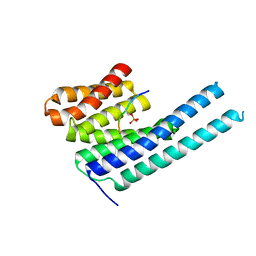 | |
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
8DGO
 
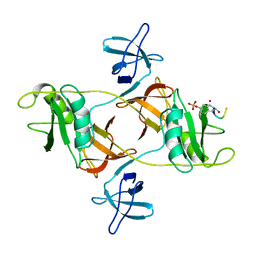 | |
8DGP
 
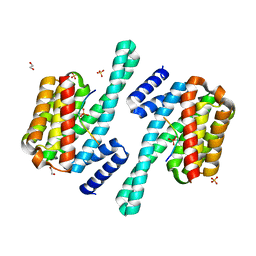 | | 14-3-3 epsilon bound to phosphorylated PEAK3 (pS69) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Phosphorylated PEAK3 (pS69) peptide, ... | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
