7RUC
 
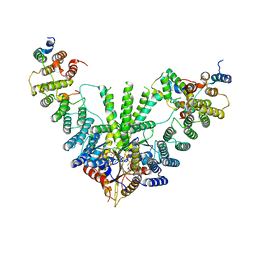 | | Metazoan pre-targeting GET complex with SGTA (cBUGGS) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 4 homolog, ... | | Authors: | Keszei, A.F.A, Yip, M.C.J, Shao, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into metazoan pretargeting GET complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7RU9
 
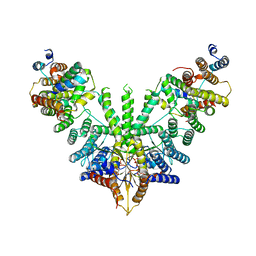 | | Metazoan pre-targeting GET complex (cBUGG-in) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 4 homolog, ... | | Authors: | Keszei, A.F.A, Yip, M.C.J, Shao, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into metazoan pretargeting GET complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7RUA
 
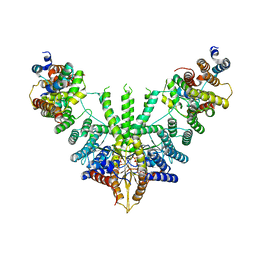 | | Metazoan pre-targeting GET complex (cBUGG-out) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 4 homolog, ... | | Authors: | Keszei, A.F.A, Yip, M.C.J, Shao, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into metazoan pretargeting GET complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MDZ
 
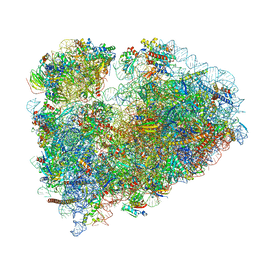 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
8T7T
 
 | |
6WT4
 
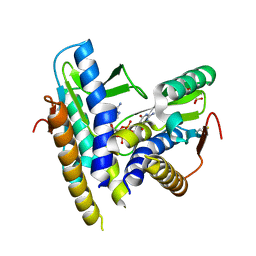 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT7
 
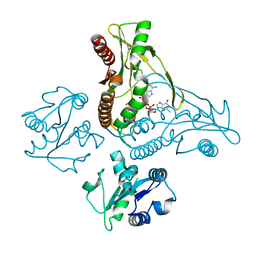 | | Structure of a metazoan TIR-STING receptor from C. gigas in complex with 2',3'-cGAMP | | Descriptor: | Metazoan TIR-STING fusion, cGAMP | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
7UN8
 
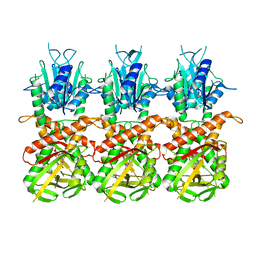 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UNA
 
 | | SfSTING with cGAMP (masked) | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
8TTQ
 
 | |
6VM6
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 2'3'3'-cAAA | | Descriptor: | 2'-5'-Linked Cyclic RNA (5'-R(P*AP*AP*A)-3'), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6WAN
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6WAM
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6WT6
 
 | | Structure of a metazoan TIR-STING receptor from C. gigas | | Descriptor: | Metazoan TIR-STING fusion | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT9
 
 | | Structure of STING-associated CdnE c-di-GMP synthase from Capnocytophaga granulosa | | Descriptor: | NTP_transf_2 domain-containing protein | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6WT5
 
 | | Structure of a bacterial STING receptor from Capnocytophaga granulosa | | Descriptor: | Bacterial STING | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6VM5
 
 | | Structure of Moraxella osloensis Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6WT8
 
 | | Structure of a STING-associated CdnE c-di-GMP synthase from Flavobacteriaceae sp. | | Descriptor: | STING-associated CdnE c-di-GMP synthase | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
8GKC
 
 | |
8EYK
 
 | |
8EYI
 
 | |
6D6I
 
 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
