3RFW
 
 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Cell-binding factor 2 | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
3RGC
 
 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Possible periplasmic protein | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3H8E
 
 | |
3H8F
 
 | | High pH native structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | BICARBONATE ION, Cytosol aminopeptidase, MANGANESE (II) ION, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
5FDK
 
 | |
4CMH
 
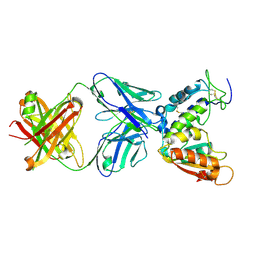 | | Crystal structure of CD38 with a novel CD38-targeting antibody SAR650984 | | Descriptor: | ADP-RIBOSYL CYCLASE 1, HEAVY CHAIN OF SAR650984-FAB FRAGMENT, LIGHT CHAIN OF SAR650984-FAB FRAGMENT | | Authors: | Deckert, J, Wetzel, M.C, Park, P.U, Bartle, L.M, Skaletskaya, A, Goldmacher, V, Vallee, F, ZhouLiu, Q, Ferrari, P, Pouzieux, S, Lahoute, C, Dumontet, C, Plesa, A, Chiron, M, Lejeune, P, Chittenden, T, Blanc, V. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SAR650984, a novel humanized CD38-targeting antibody, demonstrates potent antitumor activity in models of multiple myeloma and other CD38+ hematologic malignancies.
Clin. Cancer Res., 20, 2014
|
|
8DGZ
 
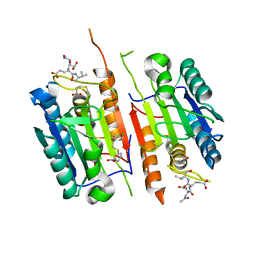 | | Caspase-7 bound to substrate mimic and allosteric inhibitor | | Descriptor: | 2-{[2-(4-chlorophenyl)-2-oxoethyl]sulfanyl}benzoic acid, Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Propp, J, Kalenkiewicz, A, Kathryn, F.H, Spies, M.A. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power.
Chemistry, 29, 2023
|
|
8DJ3
 
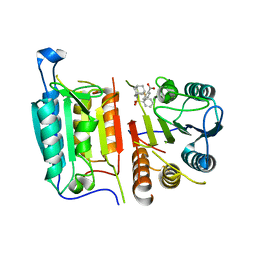 | | Caspase-7 bound to novel allosteric inhibitor | | Descriptor: | 2-[(2-{[(3s,5s,7s)-adamantan-1-yl]sulfamoyl}phenyl)sulfanyl]benzoic acid, Caspase-7 | | Authors: | Propp, J, Kalenkiewicz, A, Kathryn, F.H, Spies, M.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power.
Chemistry, 29, 2023
|
|
3NBU
 
 | | Crystal structure of pGI glucosephosphate isomerase | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase | | Authors: | Alber, T, Zubieta, C, Totir, M, May, A, Echols, N. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
6EIO
 
 | | Crystal structure of an ice binding protein from an Antarctic Biological Consortium | | Descriptor: | Antifreeze protein, GLYCEROL, SULFATE ION | | Authors: | Nardini, M, Mangiagalli, M, Nardone, V, Bar Dolev, M, Vena, V.F, Sarusi, G, Braslavsky, I, Lotti, M. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Structure of a bacterial ice binding protein with two faces of interaction with ice.
FEBS J., 285, 2018
|
|
2XHY
 
 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
5E12
 
 | |
5E10
 
 | |
5E0Y
 
 | |
5E0Z
 
 | |
3OUV
 
 | |
3SBO
 
 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
