4UBS
 
 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2CZQ
 
 | | A novel cutinase-like protein from Cryptococcus sp. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, cutinase-like protein | | Authors: | Masaki, K, Kamini, N.R, Ikeda, H, Iefuji, H, Kondo, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-14 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Proteins, 77, 2009
|
|
2RDR
 
 | | Crystal Structure of PtlH with Fe/oxalylglycine bound | | Descriptor: | 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, FE (III) ION, MAGNESIUM ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
2RDQ
 
 | | Crystal Structure of PtlH with Fe/alpha ketoglutarate bound | | Descriptor: | 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
2RDS
 
 | | Crystal Structure of PtlH with Fe/oxalylglycine and ent-1-deoxypentalenic acid bound | | Descriptor: | (1S,3aS,5aR,8aS)-1,7,7-trimethyl-1,2,3,3a,5a,6,7,8-octahydrocyclopenta[c]pentalene-4-carboxylic acid, 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, FE (III) ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
2RDN
 
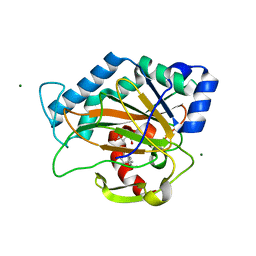 | | Crystal Structure of PtlH with AKG and ent-1PL bound | | Descriptor: | (1S,3aS,5aR,8aS)-1,7,7-trimethyl-1,2,3,3a,5a,6,7,8-octahydrocyclopenta[c]pentalene-4-carboxylic acid, 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, 2-OXOGLUTARIC ACID, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
3E5J
 
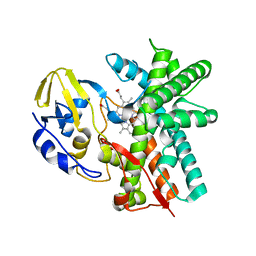 | | Crystal structure of CYP105P1 wild-type ligand-free form | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3E5K
 
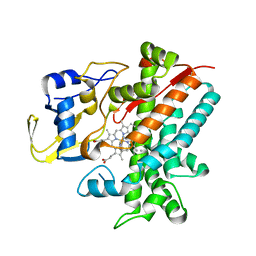 | | Crystal structure of CYP105P1 wild-type 4-phenylimidazole complex | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3E5L
 
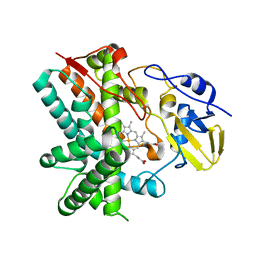 | | Crystal structure of CYP105P1 H72A mutant | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3ABA
 
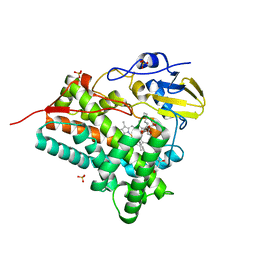 | | Crystal structure of CYP105P1 in complex with filipin I | | Descriptor: | (3R,4S,6S,8S,10R,12R,14R,16S,17E,19E,21E,23E,25E,28R)-3-hexyl-4,6,8,10,12,14,16-heptahydroxy-17,28-dimethyloxacyclooctacosa-17,19,21,23,25-pentaen-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Xu, L.H, Fushinobu, S, Takamatsu, S, Wakagi, T, Ikeda, H, Shoun, H. | | Deposit date: | 2009-12-04 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis
J.Biol.Chem., 285, 2010
|
|
3ABB
 
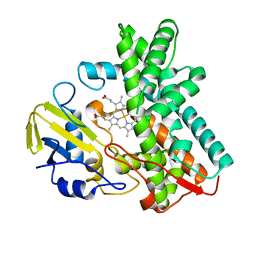 | | Crystal structure of CYP105D6 | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Takamatsu, S, Wakagi, T, Ikeda, H, Shoun, H. | | Deposit date: | 2009-12-04 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis
J.Biol.Chem., 285, 2010
|
|
388D
 
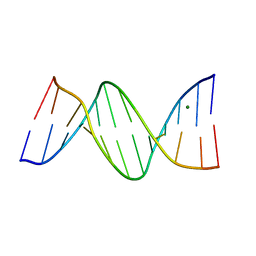 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*(TAF)P*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
389D
 
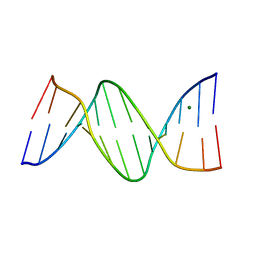 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-01-24 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
3VMM
 
 | |
