8S4T
 
 | |
8S4S
 
 | | PrgE from plasmid pCF10 | | Descriptor: | POTASSIUM ION, PrgE, SULFATE ION | | Authors: | Breidenstein, A, Berntsson, R.P.-A. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PrgE: an OB-fold protein from plasmid pCF10 with striking differences to prototypical bacterial SSBs.
Life Sci Alliance, 7, 2024
|
|
5KH1
 
 | |
8QCB
 
 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, Superkiller protein 3, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QCA
 
 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8Q9T
 
 | | CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA | | Descriptor: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QCF
 
 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
4XZX
 
 | | Shigella flexneri effector OspI C62S mutant | | Descriptor: | ACETATE ION, ORF169b | | Authors: | Nishide, A, Takagi, K, Minsoo, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New insights into the active site structure of Shigella effecter OspI
To Be Published
|
|
5H81
 
 | | HETEROYOHIMBINE SYNTHASE THAS2 FROM CATHARANTHUS ROSEUS - COMPLEX WITH NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION, heteroyohimbine synthase THAS2 | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5H82
 
 | | HETEROYOHIMBINE SYNTHASE THAS2 FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase THAS2 | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
8A1B
 
 | |
8A1C
 
 | |
5FI5
 
 | | HETEROYOHIMBINE SYNTHASE THAS1 FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | PHOSPHATE ION, Tetrahydroalstonine synthase, ZINC ION | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5FI3
 
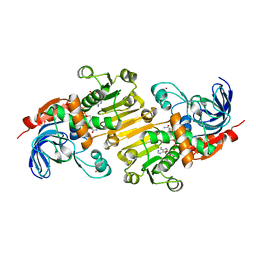 | | HETEROYOHIMBINE SYNTHASE THAS1 FROM CATHARANTHUS ROSEUS - COMPLEX WITH NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tetrahydroalstonine synthase, ... | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
3W31
 
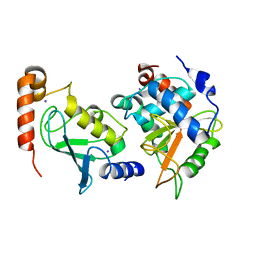 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W30
 
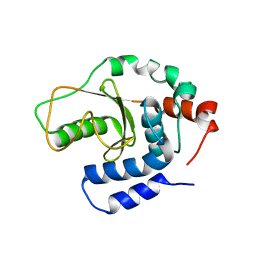 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | ORF169b | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
5A5B
 
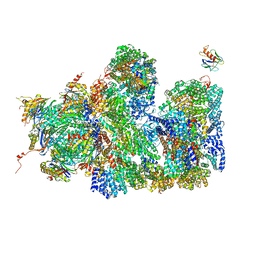 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3G9H
 
 | | Crystal structure of the C-terminal mu homology domain of Syp1 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
3G9G
 
 | | Crystal Structure of the N-terminal EFC/F-BAR domain of Syp1 | | Descriptor: | Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
3UYO
 
 | |
2P49
 
 | | Complex of a camelid single-domain vhh antibody fragment with RNASE A at 1.4A resolution: native mono_1 crystal form | | Descriptor: | ANTIBODY CAB-RN05, PHOSPHATE ION, Ribonuclease pancreatic | | Authors: | Tereshko, V, Uysal, S, Margalef, K, Koide, A, Kossiakoff, A.A, Koide, S. | | Deposit date: | 2007-03-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope.
J.Mol.Biol., 373, 2007
|
|
2P4A
 
 | | X-ray structure of a camelid affinity matured single-domain vhh antibody fragment in complex with RNASE A | | Descriptor: | ANTIBODY CAB-RN05, Ribonuclease pancreatic, SULFATE ION | | Authors: | Tereshko, V, Koide, A, Uysal, S, Koide, S. | | Deposit date: | 2007-03-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope.
J.Mol.Biol., 373, 2007
|
|
2OCF
 
 | | Human estrogen receptor alpha ligand-binding domain in complex with estradiol and the E2#23 FN3 monobody | | Descriptor: | ESTRADIOL, Estrogen receptor, Fibronectin | | Authors: | Rajan, S.S, Kuruvilla, S.M, Sharma, S.K, Kim, Y, Huang, J, Koide, A, Koide, S, Joachimiak, A, Greene, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1VDS
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
