5AYR
 
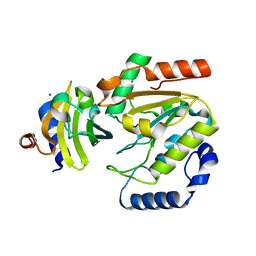 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5AYS
 
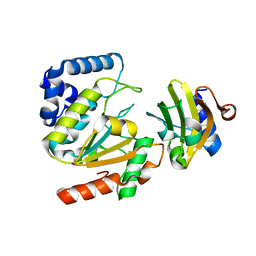 | | Crystal structure of SAUGI/HSV UDG complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5XUX
 
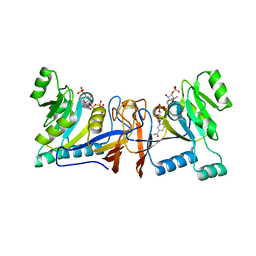 | | Crystal structure of Rib7 from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
3X0T
 
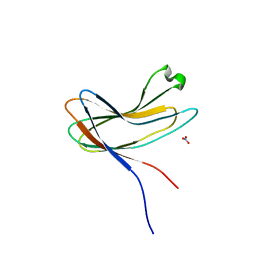 | | Crystal structure of PirA | | Descriptor: | NITRATE ION, Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3X0U
 
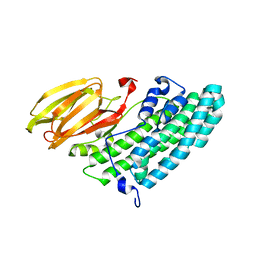 | | Crystal structure of PirB | | Descriptor: | Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8TQ2
 
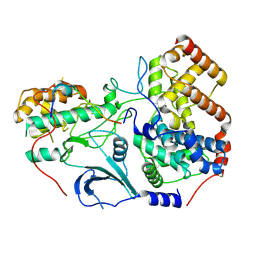 | | Structure of the kinase lobe of human CDK8 kinase module | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 12, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-06 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TQW
 
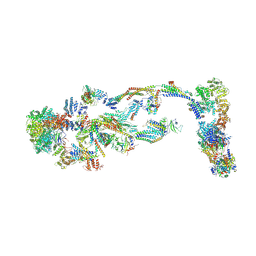 | | Structure of human transcriptional Mediator complex | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-08 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TQC
 
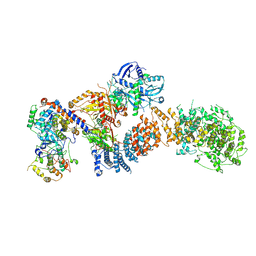 | | Structure of the human CDK8 kinase module | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 12, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-06 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TRH
 
 | | The IDRc bound human core Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
5XV2
 
 | | Crystal structure of Rib7 mutant D33A from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV0
 
 | | Crystal structure of Rib7 mutant D33N from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV5
 
 | | Crystal structure of Rib7 mutant S88E from Methanosarcina mazei | | Descriptor: | Conserved protein | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
