8SMV
 
 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
1XCT
 
 | | Complex HCV core-Fab 19D9D6-Protein L mutant (D55A, L57H, Y64W) in space group P21212 | | Descriptor: | Capsid protein C, Monoclonal antibody 19D9D6 Heavy chain, Monoclonal antibody 19D9D6 Light chain, ... | | Authors: | Menez, R, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Different crystal packing in Fab-protein L semi-disordered peptide complex.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XF5
 
 | | Complex HCV core-Fab 19D9D6-Protein L mutant (H74C, Y64W)in space group P21212 | | Descriptor: | Capsid protein C, Monoclonal antibody 19D9D6 Heavy chain, Monoclonal antibody 19D9D6 Light chain, ... | | Authors: | Menez, R, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2004-09-14 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Different crystal packing in Fab-protein L semi-disordered peptide complex.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YMH
 
 | | anti-HCV Fab 19D9D6 complexed with protein L (PpL) mutant A66W | | Descriptor: | Fab 16D9D6, heavy chain, light chain, ... | | Authors: | Granata, V, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the crystallization and crystal packing of two Fab single-site mutant protein L complexes.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XCQ
 
 | | Complex HCV core-Fab 19D9D6-Protein L mutant (D55A,L57H,Y64W) in space group P21 | | Descriptor: | Capsid protein C, Monoclonal antibody 19D9D6 Heavy chain, Monoclonal antibody 19D9D6 Light chain, ... | | Authors: | Menez, R, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Different crystal packing in Fab-protein L semi-disordered peptide complex.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2KVN
 
 | | Phi29 E-loop hairpin | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*AP*UP*UP*GP*AP*GP*UP*UP*CP*AP*CP*CP*A)-3') | | Authors: | Harris, S.M, Schroeder, S.J. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the prohead RNA E-loop hairpin.
Biochemistry, 49, 2010
|
|
2N5G
 
 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
4HVI
 
 | |
4HVH
 
 | |
4HVG
 
 | |
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
3D14
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)-ethyl]- thiazol-2-yl}-3-(3-trifluoromethyl-phenyl)-urea | | Descriptor: | 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Baskaran, S, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D15
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-(3-chloro-phenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)- ethyl]-thiazol-2-yl}-urea [SNS-314] | | Descriptor: | 1-(3-chlorophenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Bui, M, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4PCO
 
 | |
7LX2
 
 | | Cryo-EM structure of ConSOSL.UFO.664 (ConS) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LX3
 
 | | Cryo-EM structure of EDC-crosslinked ConSOSL.UFO.664 (ConS-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXM
 
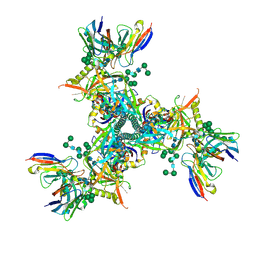 | | Cryo-EM structure of ConM SOSIP.v7 (ConM) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LXN
 
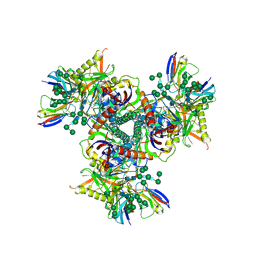 | | Cryo-EM structure of EDC-crosslinked ConM SOSIP.v7 (ConM-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env glycoprotein gp120, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
8TQD
 
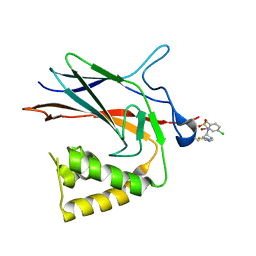 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
4WTU
 
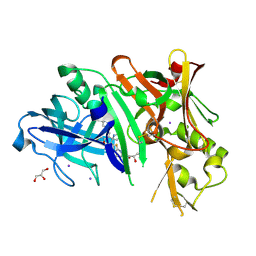 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-aza-4-fluoro-xanthene inhibitor 22 | | Descriptor: | (5S)-3-(5,6-dihydro-2H-pyran-3-yl)-1-fluoro-7-(2-fluoropyridin-3-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Orally Available BACE1 Inhibitor That Affords Robust CNS A beta Reduction without Cardiovascular Liabilities.
Acs Med.Chem.Lett., 6, 2015
|
|
7KW1
 
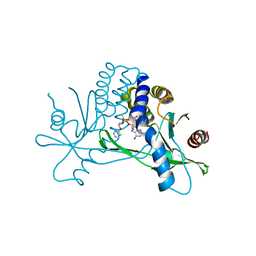 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
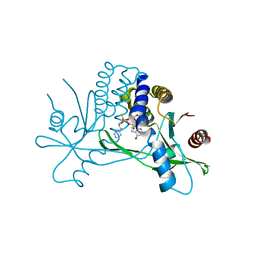 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVZ
 
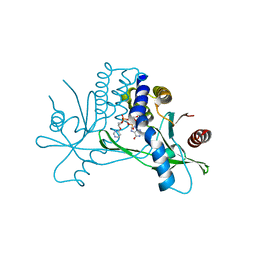 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
1MHH
 
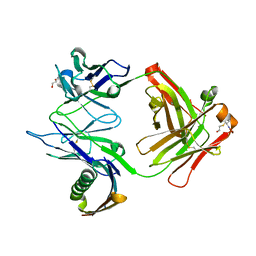 | |
1LCJ
 
 | |
