3C91
 
 | | Thermoplasma acidophilum 20S proteasome with an open gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3C92
 
 | | Thermoplasma acidophilum 20S proteasome with a closed gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
3IPM
 
 | |
5E0S
 
 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5DZK
 
 | | Crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, BEZ-LEU-LEU | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
3MDJ
 
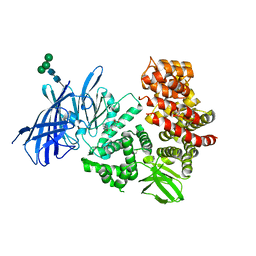 | | ER Aminopeptidase, ERAP1, Bound to the Zinc Aminopeptidase Inhibitor, Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nguyen, T.T, Stern, L.J. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6VGQ
 
 | | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, Z-Gly-leu-phe-CH2Cl | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VGK
 
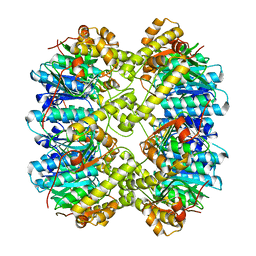 | | ClpP1P2 complex from M. tuberculosis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QXN
 
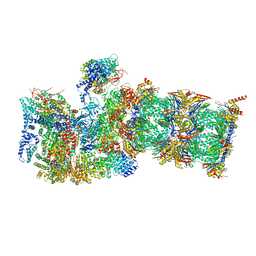 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
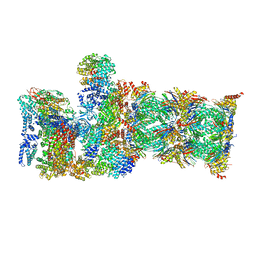 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXP
 
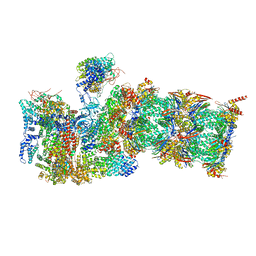 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXX
 
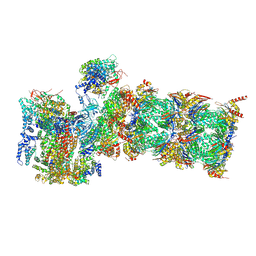 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
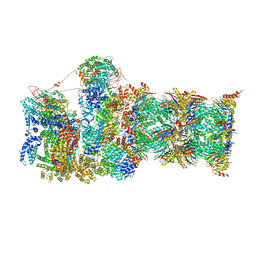 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QY7
 
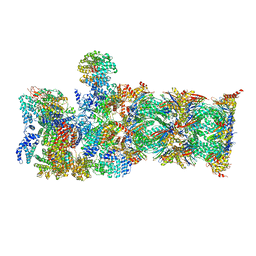 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXW
 
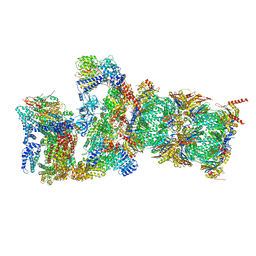 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXU
 
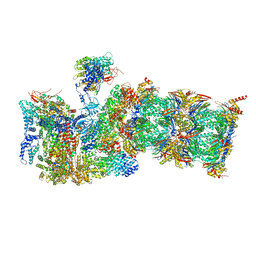 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
6VGN
 
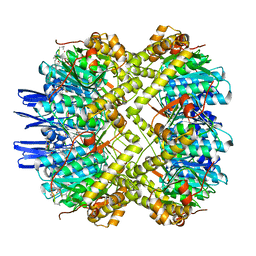 | | ClpP1P2 complex from M. tuberculosis bound to ADEP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, R0M-WFP-ALO-PRO-YCP-ALA-MP8 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2FZP
 
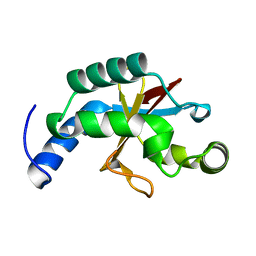 | | Crystal structure of the USP8 interaction domain of human NRDP1 | | Descriptor: | ring finger protein 41 isoform 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
1DO0
 
 | | ORTHORHOMBIC CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (HEAT SHOCK LOCUS U), ... | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
1DO2
 
 | | TRIGONAL CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (HEAT SHOCK LOCUS U) | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
