1QK9
 
 | | The solution structure of the domain from MeCP2 that binds to methylated DNA | | Descriptor: | METHYL-CPG-BINDING PROTEIN 2 | | Authors: | Wakefield, R.I.D, Smith, B.O, Nan, X, Free, A, Soteriou, A, Uhrin, D, Bird, A.P, Barlow, P.N. | | Deposit date: | 1999-07-12 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Domain from Mecp2 that Binds to Methylated DNA
J.Mol.Biol., 291, 1999
|
|
3F2L
 
 | | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Frolow, F, Freeman, A, Wine, Y, Eppel, A, Shanzer, M, Stempler, S. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus
To be Published
|
|
2HU1
 
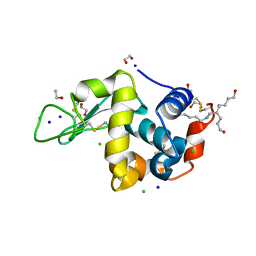 | | Crystal structure Analysis of Hen Egg White Lyszoyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wine, Y, Cohen-Hadar, N, Freeman, A, Lagziel-Simis, S, Frolow, F. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis
Biotechnol.Bioeng., 98, 2007
|
|
2HU3
 
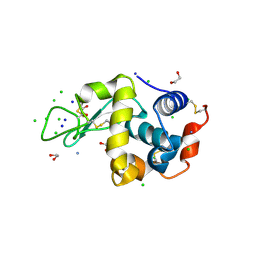 | | Parent Structure of Hen Egg White Lysozyme grown in acidic pH 4.8. Refinement for comparison with crosslinked molecules of lysozyme | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Lagziel-Simis, S, Cohen-Hadar, N, Wine, Y, Freeman, A. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Monitoring influence of pH on the molecular and crystal structure of hen egg-white tetragonal lysozyme
To be Published
|
|
2HTX
 
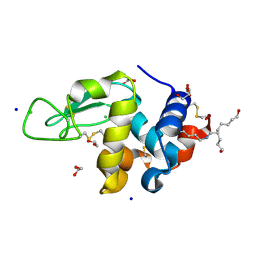 | | Crystal Structure Analysis of Hen Egg White Lysozyme Crosslinked by Polymerized Glutaraldehyde in Acidic Environment | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wine, Y, Cohen-Hadar, N, Freeman, A, Lagziel-Simis, S, Frolow, F. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis
Biotechnol.Bioeng., 98, 2007
|
|
2HUB
 
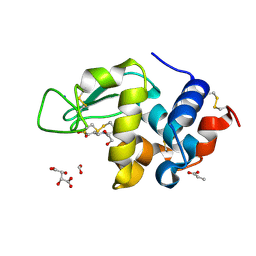 | | Structure of Hen Egg-White Lysozyme Determined from crystals grown in pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D(-)-TARTARIC ACID, ... | | Authors: | Frolow, F, Lagziel-Simis, S, Cohen-Hadar, N, Wine, Y, Freeman, A. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Monitoring influence of pH on the molecular and crystal structure of hen egg-white tetragonal lysozyme
To be Published
|
|
6PXO
 
 | |
1KZU
 
 | | INTEGRAL MEMBRANE PERIPHERAL LIGHT HARVESTING COMPLEX FROM RHODOPSEUDOMONAS ACIDOPHILA STRAIN 10050 | | Descriptor: | BACTERIOCHLOROPHYLL A, LIGHT HARVESTING PROTEIN B-800/850, Rhodopin b-D-glucoside | | Authors: | Cogdell, R.J, Freer, A.A, Isaacs, N.W, Hawthornthwaite-Lawless, A.M, Mcdermott, G, Papiz, M.Z, Prince, S.M. | | Deposit date: | 1996-08-31 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophila strain 10050: modular assembly and protein pigment interactions.
J.Mol.Biol., 268, 1997
|
|
2XYA
 
 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3DOK
 
 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | Descriptor: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DLE
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DOL
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DLG
 
 | | Crystal structure of hiv-1 reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, P51 RT, PHOSPHATE ION, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
