1WSX
 
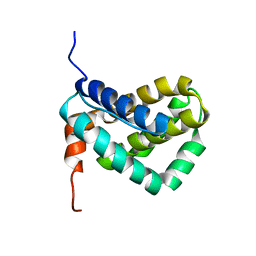 | | Solution structure of MCL-1 | | Descriptor: | myeloid cell leukemia sequence 1 | | Authors: | Day, C.L, Chen, L, Richardson, S.J, Harrison, P.J, Huang, D.C, Hinds, M.G. | | Deposit date: | 2004-11-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Prosurvival Mcl-1 and Characterization of Its Binding by Proapoptotic BH3-only Ligands
J.Biol.Chem., 280, 2005
|
|
1RE6
 
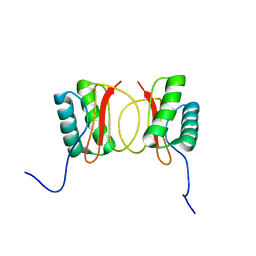 | | Localisation of Dynein Light Chains 1 and 2 and their Pro-apoptotic Ligands | | Descriptor: | dynein light chain 2 | | Authors: | Day, C.L, Puthalakath, H, Skea, G, Strasser, A, Barsukov, I, Lian, L.Y, Huang, D.C, Hinds, M.G. | | Deposit date: | 2003-11-06 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands.
Biochem.J., 377, 2004
|
|
1DEB
 
 | |
1CWW
 
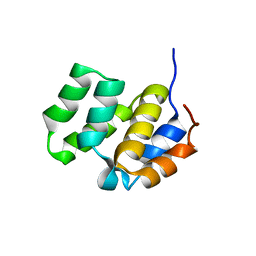 | | SOLUTION STRUCTURE OF THE CASPASE RECRUITMENT DOMAIN (CARD) FROM APAF-1 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Day, C.L, Dupont, C, Lackmann, M, Vaux, D.L, Hinds, M.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-01-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1.
Cell Death Differ., 6, 1999
|
|
2ROC
 
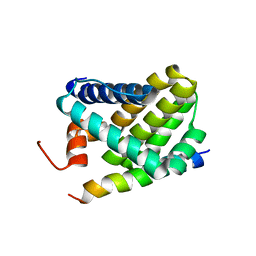 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2ROD
 
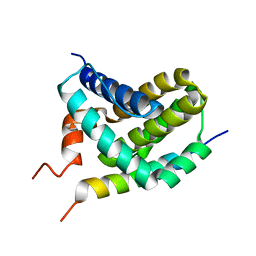 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
1LCT
 
 | |
2L9M
 
 | | Structure of cIAP1 CARD | | Descriptor: | Baculoviral IAP repeat-containing protein 2 | | Authors: | Day, C.L, Rautureau, G.J.P, Hinds, M.G. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration.
Mol.Cell, 42, 2011
|
|
9N1F
 
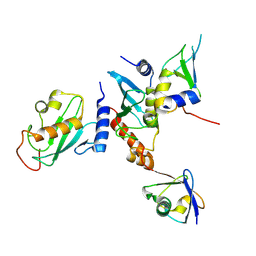 | | Crystal Structure of the Ark2C-Ubc13~Ub-Mms2 complex | | Descriptor: | E3 ubiquitin-protein ligase ARK2C, Ubiquitin, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Paluda, A, Middleton, A.J, Day, C.L. | | Deposit date: | 2025-01-26 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arkadia and Ark2C Promote Substrate Ubiquitylation with Multiple E2 Enzymes.
J.Mol.Biol., 437, 2025
|
|
5D0K
 
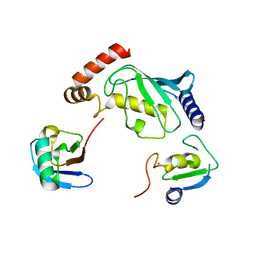 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | Polyubiquitin-B, RING finger protein 165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D0I
 
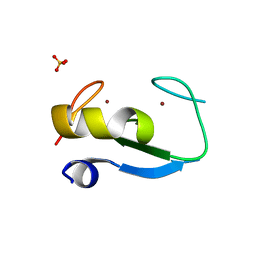 | | Structure of RING finger protein 165 | | Descriptor: | RING finger protein 165, SULFATE ION, ZINC ION | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D0M
 
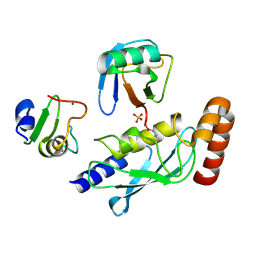 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | PHOSPHATE ION, Polyubiquitin-B, RING finger protein 165, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DFL
 
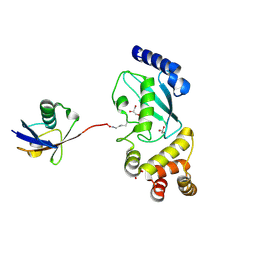 | |
2VJF
 
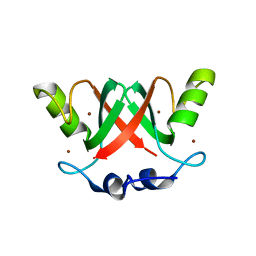 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mdm2/Mdmx Ring Domain Heterodimer Reveals Dimerization is Required for Their Ubiquitylation in Trans.
Cell Death Differ., 15, 2008
|
|
2VJE
 
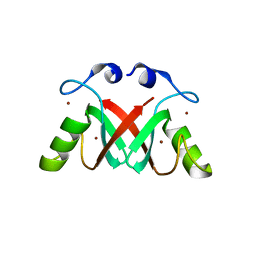 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans.
Cell Death Differ., 15, 2008
|
|
1VFD
 
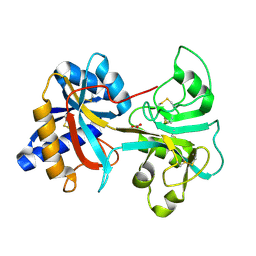 | | HUMAN LACTOFERRIN, N-TERMINAL LOBE MUTANT WITH ARG 121 REPLACED BY GLU (R121E) | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Faber, H.R, Day, C.L, Baker, E.N. | | Deposit date: | 1996-10-01 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of arginine 121 in lactoferrin destabilizes iron binding by disruption of anion binding: crystal structures of R121S and R121E mutants.
Biochemistry, 35, 1996
|
|
1VFE
 
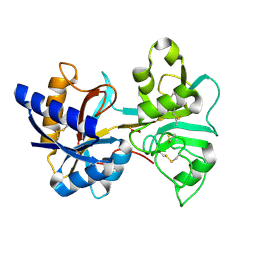 | | HUMAN LACTOFERRIN, N-TERMINAL LOBE MUTANT WITH ARG 121 REPLACED BY SER (R121S) | | Descriptor: | CARBONATE ION, FE (III) ION, HUMAN LACTOFERRIN | | Authors: | Faber, H.R, Day, C.L, Baker, E.N. | | Deposit date: | 1996-10-01 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutation of arginine 121 in lactoferrin destabilizes iron binding by disruption of anion binding: crystal structures of R121S and R121E mutants.
Biochemistry, 35, 1996
|
|
5TRB
 
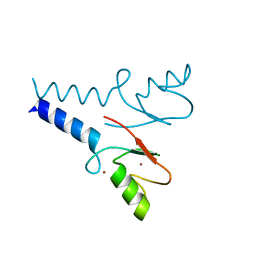 | | Crystal structure of the RNF20 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1A, ZINC ION | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B.
J.Mol.Biol., 428, 2016
|
|
8GCB
 
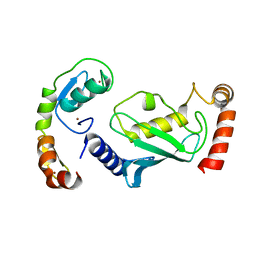 | | Structure of RNF125 in complex with a UbcH5b~Ub conjugate | | Descriptor: | E3 ubiquitin-protein ligase RNF125, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
8GBQ
 
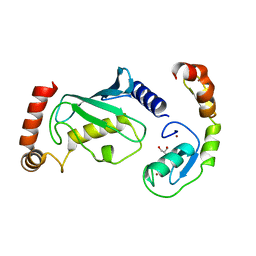 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
7R70
 
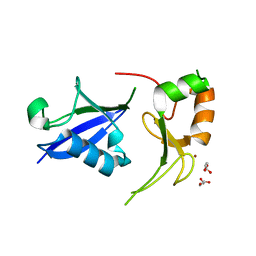 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
7R71
 
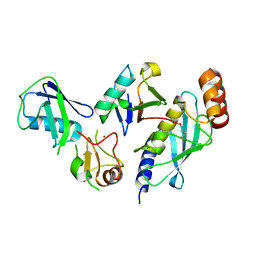 | | Crystal Structure of the UbArk2C-UbcH5b~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin,E3 ubiquitin-protein ligase RNF165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
3EB6
 
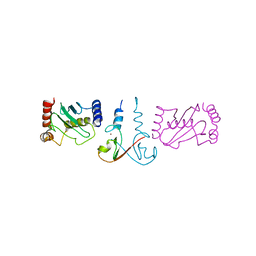 | | Structure of the cIAP2 RING domain bound to UbcH5b | | Descriptor: | Baculoviral IAP repeat-containing protein 3, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Mace, P.D, Linke, K, Schumacher, F.-R, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment.
J.Biol.Chem., 283, 2008
|
|
3EB5
 
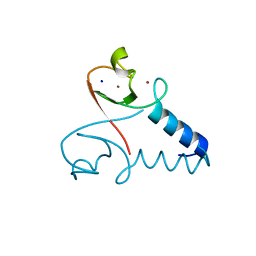 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
4IC2
 
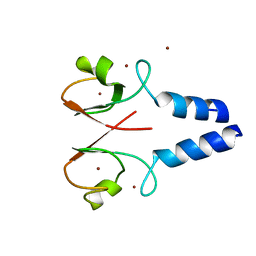 | | Crystal structure of the XIAP RING domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, NICKEL (II) ION, ZINC ION | | Authors: | Linke, K, Nakatani, Y, Day, C.L. | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase
Biochem.J., 450, 2013
|
|
