2N0Y
 
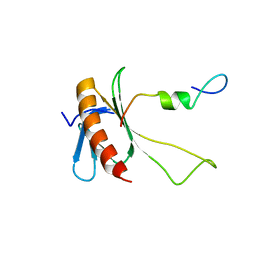 | | NMR structure of the complex between the C-terminal domain of the Rift Valley fever virus protein NSs and the PH domain of the Tfb1 subunit of TFIIH | | Descriptor: | Non-structural protein NS-S, RNA polymerase II transcription factor B subunit 1 | | Authors: | Cyr, N, de la Fuente, C, Lecoq, L, Guendel, I, Chabot, P.R, Kehn-Hall, K, Omichinski, J.G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Omega XaV motif in the Rift Valley fever virus NSs protein is essential for degrading p62, forming nuclear filaments and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8T2N
 
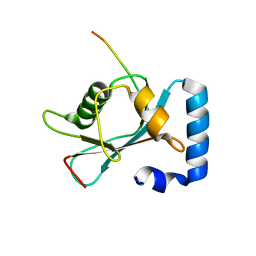 | | Crystal structure of GABARAP in complex with the LIR of NSs3 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Non-structural protein S | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2L
 
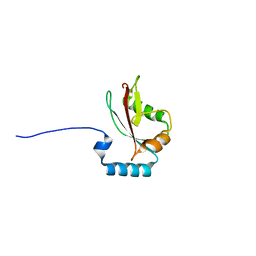 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2M
 
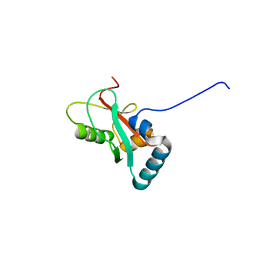 | | Crystal structure of GABARAP in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Gamma-aminobutyric acid receptor-associated protein chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
5U4K
 
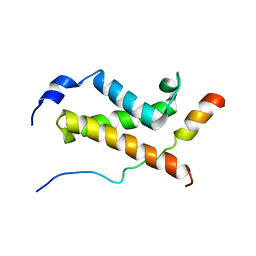 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
5URN
 
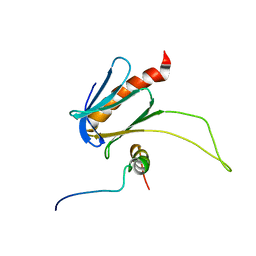 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
8T32
 
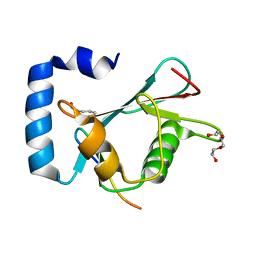 | | Crystal structure of K48 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, LIR of DOR, TRIETHYLENE GLYCOL | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T31
 
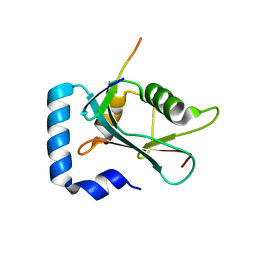 | | Crystal structure of GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2 | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T35
 
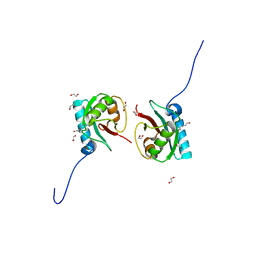 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T4T
 
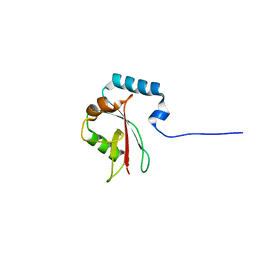 | | Crystal structure of LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T36
 
 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T33
 
 | | Crystal structure of K46 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | ACETATE ION, Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2, ... | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
6U0P
 
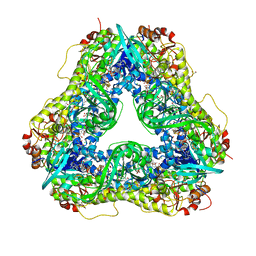 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6U0S
 
 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6HKU
 
 | |
6HKV
 
 | |
