5CCA
 
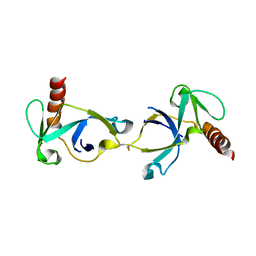 | | Crystal structure of Mtb toxin | | Descriptor: | Endoribonuclease MazF3 | | Authors: | Cascio, D, Arbing, M, de Serrano, V, Eisenberg, D, Miallau, L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Mtb toxin
To Be Published
|
|
5HRY
 
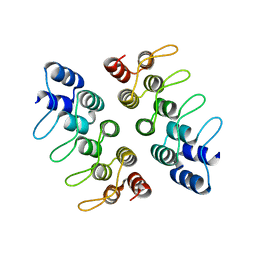 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRZ
 
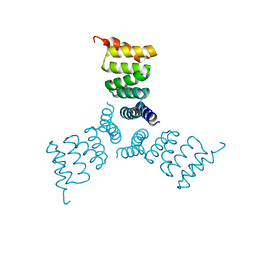 | | Computationally Designed Trimer 1na0C3_3 | | Descriptor: | TPR domain protein 1na0C3_3 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
4U9P
 
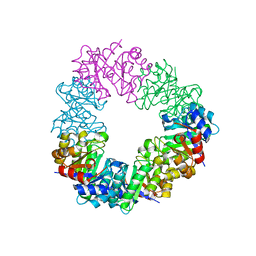 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | Authors: | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-26 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
3JRG
 
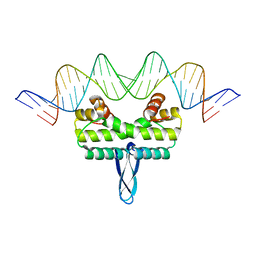 | |
3JRE
 
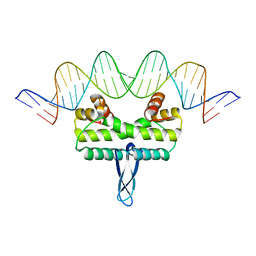 | |
5K7P
 
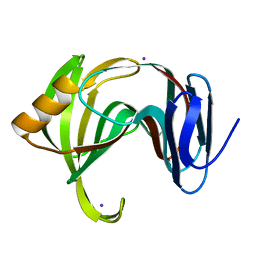 | | MicroED structure of xylanase at 2.3 A resolution | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
3RUB
 
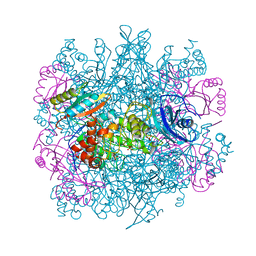 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
2IBP
 
 | |
2IEF
 
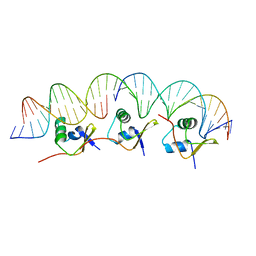 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1COS
 
 | | CRYSTAL STRUCTURE OF A SYNTHETIC TRIPLE-STRANDED ALPHA-HELICAL BUNDLE | | Descriptor: | COILED SERINE | | Authors: | Lovejoy, B, Choe, S, Cascio, D, Mcrorie, D.K, Degrado, W, Eisenberg, D. | | Deposit date: | 1993-01-22 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a synthetic triple-stranded alpha-helical bundle.
Science, 259, 1993
|
|
5K2H
 
 | |
1YAC
 
 | |
4D9J
 
 | |
1RLC
 
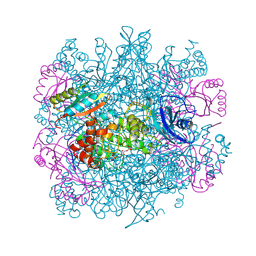 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
6E72
 
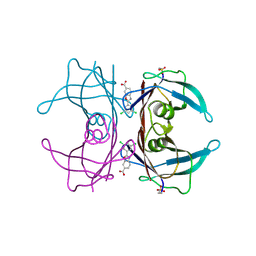 | | Structure of Human Transthyretin Val30Met Mutant in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E71
 
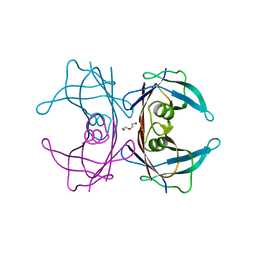 | | Structure of Human Transthyretin Val30Met/Thr119Met Mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Transthyretin | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E78
 
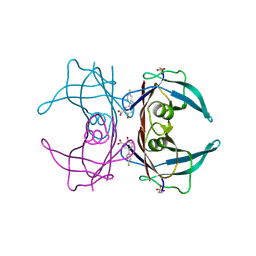 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
1BYZ
 
 | | DESIGNED PEPTIDE ALPHA-1, P1 FORM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Prive, G.G, Anderson, D.H, Wesson, L, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-20 | | Release date: | 1998-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Packed protein bilayers in the 0.90 A resolution structure of a designed alpha helical bundle.
Protein Sci., 8, 1999
|
|
4UNT
 
 | | Induced monomer of the Mcg variable domain | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C.M, Whitelegge, J.P, Phillips, M.L, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
1ZWN
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1B) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-03 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1B)
To be Published
|
|
1ZUR
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1F) | | Descriptor: | CHLORIDE ION, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-4-phenyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-05-31 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1F)
To be Published
|
|
2A4T
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R7) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-29 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R7
To be Published
|
|
1ZYT
 
 | | Crystal structure of spin labeled T4 Lysozyme (A82R1) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | EPR (1.7 Å), X-RAY DIFFRACTION | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins
Protein Sci., 18, 2009
|
|
