1RJB
 
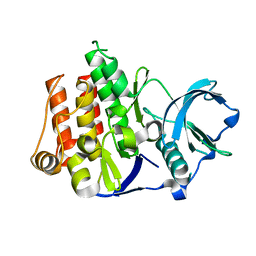 | | Crystal Structure of FLT3 | | Descriptor: | FL cytokine receptor | | Authors: | Griffith, J, Black, J, Faerman, C, Swenson, L, Wynn, M, Lu, F, Lippke, J, Saxena, K. | | Deposit date: | 2003-11-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for autoinhibition of FLT3 by the juxtamembrane domain.
Mol.Cell, 13, 2004
|
|
1YHS
 
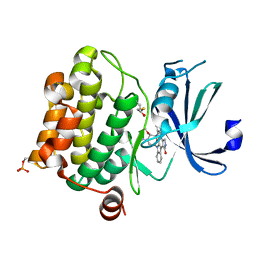 | | Crystal structure of Pim-1 bound to staurosporine | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1, STAUROSPORINE | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-10 | | Release date: | 2005-01-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1YI3
 
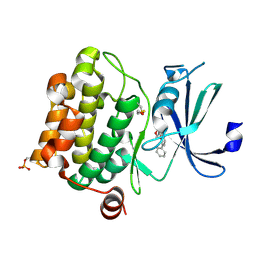 | | Crystal Structure of Pim-1 bound to LY294002 | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1YI4
 
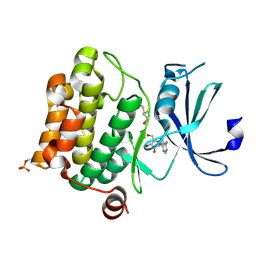 | | Structure of Pim-1 bound to adenosine | | Descriptor: | ADENOSINE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
3F71
 
 | |
3EZG
 
 | |
6O5W
 
 | | Crystal structure of MORC3 CW domain fused with viral influenza A NS1 peptide | | Descriptor: | NS1-linked peptide,MORC family CW-type zinc finger protein 3, ZINC ION | | Authors: | Ahn, J, Zhang, Y, Vann, K.R, Kutateleadze, T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | MORC3 Is a Target of the Influenza A Viral Protein NS1.
Structure, 27, 2019
|
|
6O1E
 
 | | The crystal structure of human MORC3 ATPase-CW in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mechanism for autoinhibition and activation of the MORC3 ATPase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
