1NX9
 
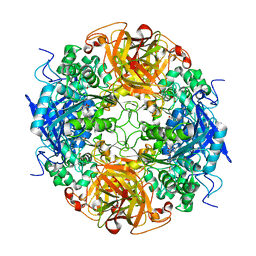 | | Acetobacter turbidans alpha-amino acid ester hydrolase S205A mutant complexed with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acetobacter turbidans {alpha}-Amino Acid Ester Hydrolase: HOW A SINGLE MUTATION IMPROVES AN ANTIBIOTIC-PRODUCING ENZYME.
J.Biol.Chem., 281, 2006
|
|
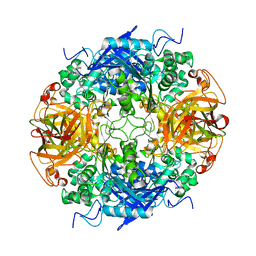
1RYY
 
 | |
1TZ7
 
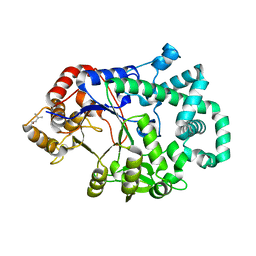 | | Aquifex aeolicus amylomaltase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-alpha-glucanotransferase | | Authors: | Barends, T.R.M, Korf, H, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2004-07-09 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural influences on product specificity in amylomaltase from Aquifex aeolicus
TO BE PUBLISHED
|
|
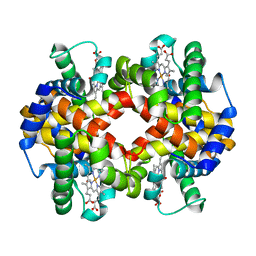
7AET
 
 | |
7AEU
 
 | |
7AEV
 
 | |
3L6W
 
 | | Structure of the collar functional unit (KLH1-H) of keyhole limpet hemocyanin | | Descriptor: | Hemocyanin 1 | | Authors: | Jaenicke, E, Buechler, K, Markl, J, Decker, H, Barends, T.R.M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Cupredoxin-like Domains in Hemocyanins.
Biochem.J., 2009
|
|
6HAL
 
 | | Human carbonmonoxy hemoglobin SFX dataset | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Doak, B, Gorel, A, Foucar, L, Barends, T.R.M, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C.M, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrell, D.A, Owen, R.L, Schlichting, I. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6HIF
 
 | | Kuenenia stuttgartiensis hydrazine dehydrogenase complex | | Descriptor: | GLYCEROL, HEME C, Hydrazine dehydrogenase, ... | | Authors: | Akram, M, Dietl, A, Mersdorf, U, Prinz, S, Maalcke, W, Keltjens, J, Ferousi, C, de Almeida, N.M, Reimann, J, Kartal, B, Jetten, M.S.M, Parey, K, Barends, T.R.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 192-heme electron transfer network in the hydrazine dehydrogenase complex.
Sci Adv, 5, 2019
|
|
9G2J
 
 | | Thaumatin structure determined using SoS chip at ID29 (serial crystallography) | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Doak, R.B, Shoeman, R.L, Gorel, A, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2024-07-11 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Sheet-on-sheet fixed target data collection devices for serial crystallography at synchrotron and XFEL sources.
J.Appl.Crystallogr., 57, 2024
|
|
9FBK
 
 | |
6T39
 
 | | Crystal structure of rsEGFP2 in its off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
8AYB
 
 | | anammox-specific FabZ from Scalindua brodae | | Descriptor: | Beta-hydroxyacyl-(Acyl-carrier-protein) dehydratase | | Authors: | Dietl, A, Barends, T.R.M. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of an unusual 3-hydroxyacyl dehydratase (FabZ) from a ladderane-producing organism with an unexpected substrate preference.
J.Biol.Chem., 299, 2023
|
|
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
2PI8
 
 | | Crystal structure of E. coli MltA with bound chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase A, PHOSPHATE ION | | Authors: | van Straaten, K.E, Barends, T.R.M, Dijkstra, B.W, Thunnissen, A.M.W.H. | | Deposit date: | 2007-04-13 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Escherichia coli Lytic transglycosylase MltA with bound chitohexaose: implications for peptidoglycan binding and cleavage
J.Biol.Chem., 282, 2007
|
|
4LJ8
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJ4
 
 | | ClpB NBD2 from T. thermophilus, nucleotide-free | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJ9
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP | | Descriptor: | Chaperone protein ClpB, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJ7
 
 | | ClpB NBD2 K601Q from T. thermophilus in complex with MANT-dADP | | Descriptor: | 2'(3')-O-N-METHYLANTHRANILOYL-ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
4LJ6
 
 | | ClpB NBD2 from T. thermophilus in complex with AMPPCP | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJA
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP and guanidinium chloride | | Descriptor: | Chaperone protein ClpB, GUANIDINE, MAGNESIUM ION, ... | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8C
 
 | | Composite structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
