3GQ1
 
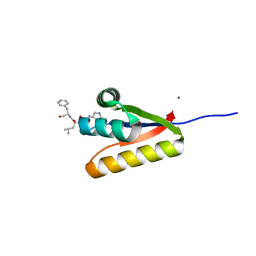 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G3P
 
 | |
3G1B
 
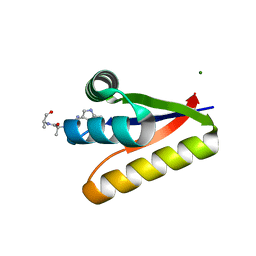 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | Descriptor: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G19
 
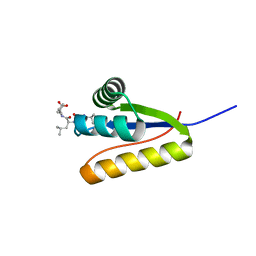 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GQ0
 
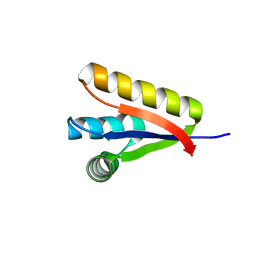 | |
3GW1
 
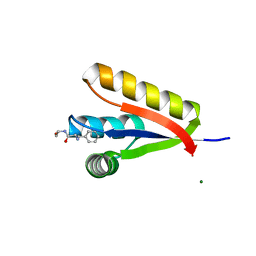 | | The structure of the Caulobacter crescentus CLPs protease adaptor protein in complex with FGG tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, FGG peptide, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1TWB
 
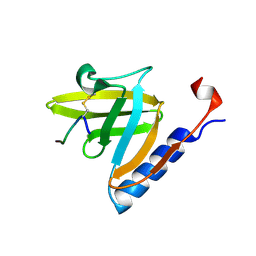 | | SspB disulfide crosslinked to an ssrA degradation tag | | Descriptor: | Stringent starvation protein B homolog, ssrA peptide | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
3O1F
 
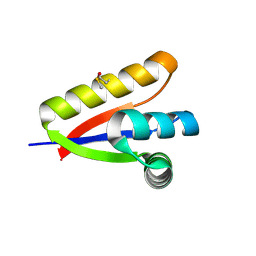 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | Descriptor: | ATP-dependent Clp protease adapter protein clpS | | Authors: | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
1YFN
 
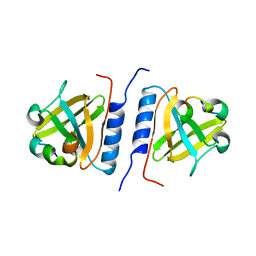 | | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB- the crystal structure of a SspB-RseA complex | | Descriptor: | Sigma-E factor negative regulatory protein, Stringent starvation protein B | | Authors: | Levchenko, I, Grant, R.A, Flynn, J.M, Sauer, R.T, Baker, T.A. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB
Nat.Struct.Mol.Biol., 12, 2005
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
1ZSZ
 
 | | Crystal structure of a computationally designed SspB heterodimer | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2005-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity versus stability in computational protein design.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3F7A
 
 | |
3HTE
 
 | | Crystal structure of nucleotide-free hexameric ClpX | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SULFATE ION | | Authors: | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.026 Å) | | Cite: | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HWS
 
 | | Crystal structure of nucleotide-bound hexameric ClpX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX, MAGNESIUM ION, ... | | Authors: | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
3F79
 
 | |
7UIY
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
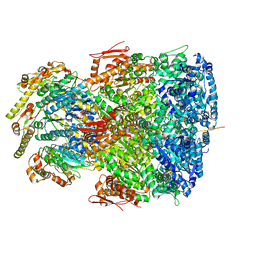 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
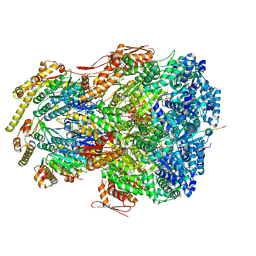 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
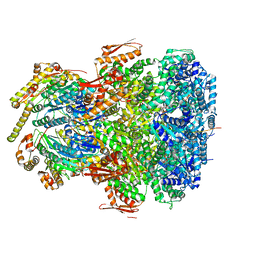 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
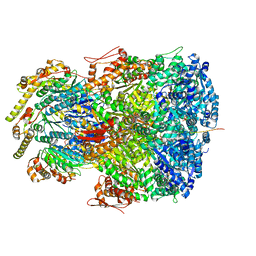 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TXT
 
 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5TXR
 
 | | Structure of ALAS from S. cerevisiae non-covalently bound to PLP cofactor | | Descriptor: | 5-aminolevulinate synthase, mitochondrial, FORMIC ACID, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
