2A6E
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A69
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
1SMY
 
 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
2PPB
 
 | | Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I, Landick, R. | | Deposit date: | 2007-04-28 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate loading in bacterial RNA polymerase.
Nature, 448, 2007
|
|
5OND
 
 | | RfaH from Escherichia coli in complex with ops DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*TP*AP*GP*TP*C)-3'), Transcription antitermination protein RfaH | | Authors: | Zuber, P.K, Artsimovitch, I, Roesch, P, Knauer, S.H. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand.
Elife, 7, 2018
|
|
3EQL
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transcription inactivation through local refolding of the RNA polymerase structure.
Nature, 457, 2009
|
|
4IJJ
 
 | | Structure of transcription factor DksA2 from Pseudomonas aeruginosa | | Descriptor: | Putative C4-type zinc finger protein, DksA/TraR family, SULFATE ION | | Authors: | Biswas, T, Furman, R, Artsimovitch, I, Tsodikov, O.V. | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-27 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DksA2, a zinc-independent structural analog of the transcription factor DksA.
Febs Lett., 587, 2013
|
|
2BE5
 
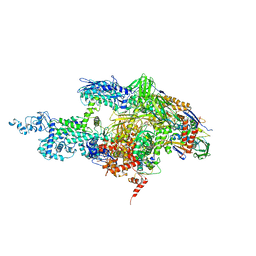 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
2EUL
 
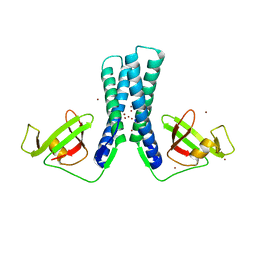 | | Structure of the transcription factor Gfh1. | | Descriptor: | ZINC ION, anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Symersky, J, Perederina, A, Vassylyeva, M.N, Svetlov, V, Artsimovitch, I, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation through the RNA Polymerase Secondary Channel: STRUCTURAL AND FUNCTIONAL VARIABILITY OF THE COILED-COIL TRANSCRIPTION FACTORS.
J.Biol.Chem., 281, 2006
|
|
1TJL
 
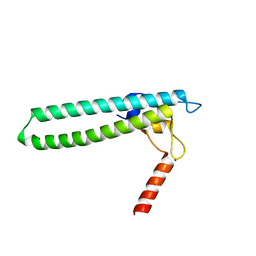 | | Crystal structure of transcription factor DksA from E. coli | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Perederina, A, Svetlov, V, Vassylyeva, M.N, Artsimovitch, I, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription
Cell(Cambridge,Mass.), 118, 2004
|
|
6C6S
 
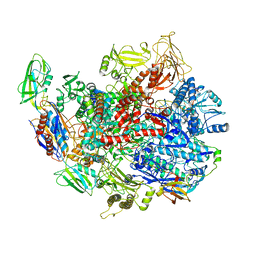 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6U
 
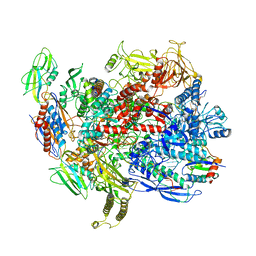 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6T
 
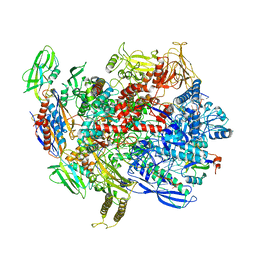 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
2OUG
 
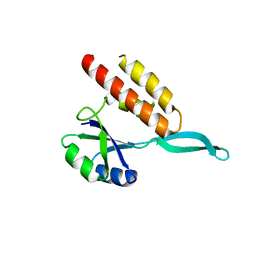 | |
2P4V
 
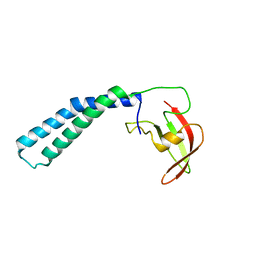 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
4DFC
 
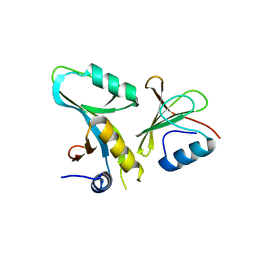 | | Core UvrA/TRCF complex | | Descriptor: | Transcription-repair-coupling factor, UvrABC system protein A | | Authors: | Deaconescu, A.M, Grigorieff, N. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TMC
 
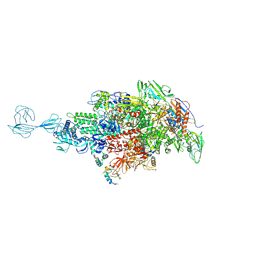 | | Re-refinement of Thermus thermopiles DNA-directed RNA polymerase structure | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
6DUQ
 
 | | Structure of a Rho-NusG KOW domain complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Berger, J.M, Lawson, M.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein.
Mol. Cell, 71, 2018
|
|
7ADD
 
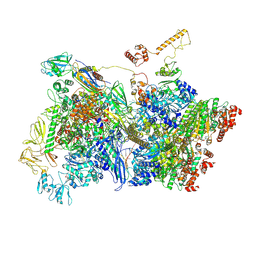 | | Transcription termination intermediate complex IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADC
 
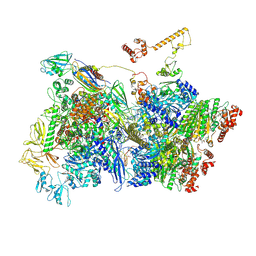 | | Transcription termination intermediate complex 3 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADE
 
 | | Transcription termination complex IVa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADB
 
 | | Transcription termination intermediate complex 1 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
2O5J
 
 | |
2O5I
 
 | | Crystal structure of the T. thermophilus RNA polymerase elongation complex | | Descriptor: | 5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3', 5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*GP*AP*GP*UP*CP*UP*GP*CP*GP*GP*CP*GP*CP*GP*CP*G)-3', ... | | Authors: | Vassylyev, D.G, Tahirov, T.H, Vassylyeva, M.N. | | Deposit date: | 2006-12-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for transcription elongation by bacterial RNA polymerase.
Nature, 448, 2007
|
|
