6M02
 
 | | cryo-EM structure of human Pannexin 1 channel | | Descriptor: | Pannexin-1 | | Authors: | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-25 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
5WE6
 
 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
7YDQ
 
 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8J17
 
 | | Crystal structure of IsPETase variant | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Yin, Q.D, Wang, Y.X. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Efficient polyethylene terephthalate biodegradation by an engineered Ideonella sakaiensis PETase with a fixed substrate-binding W156 residue
Green Chem, 2013
|
|
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7D54
 
 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5YJG
 
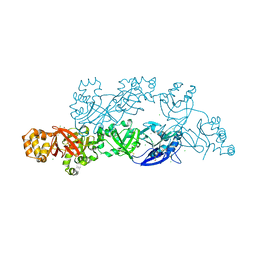 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
7XPQ
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
3K9W
 
 | |
4GND
 
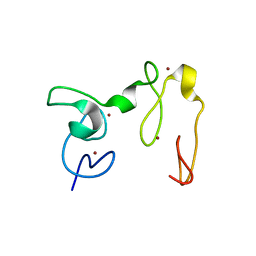 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | Descriptor: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
7XPP
 
 | |
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2017-06-24 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
6G97
 
 | |
6G9H
 
 | |
5LZ6
 
 | |
6G93
 
 | |
6G9J
 
 | |
7LDG
 
 | | Crystal structure of the MEILB2-BRCA2 complex | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein | | Authors: | Nandakumar, J, Pendlebury, D.F. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of a meiosis-specific complex central to BRCA2 localization at recombination sites.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3K9X
 
 | | X-ray crystal structure of human fxa in complex with (S)-N-((2-METHYLBENZOFURAN-5-YLAMINO)(2-OXO-1-(2-OXO-2- (PYRROLIDIN-1-YL)ETHYL)AZEPAN-3- YLAMINO)METHYLENE)NICOTINAMIDE | | Descriptor: | CALCIUM ION, GLYCEROL, N-{N'-(2-methyl-1-benzofuran-5-yl)-N-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]carbamimidoyl}pyridine-3-carboxamide, ... | | Authors: | Klei, H.E, Kish, K, Ghosh, K, Rushith, A. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aroylguanidine-based factor Xa inhibitors: the discovery of BMS-344577
Bioorg.Med.Chem.Lett., 19, 2009
|
|
