7X1Y
 
 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
8K75
 
 | |
8J5D
 
 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
8Z61
 
 | | Human beta-catenin crystal structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel 1-Phenylpiperidine Urea-Containing Derivatives Inhibiting beta-Catenin/BCL9 Interaction and Exerting Antitumor Efficacy through the Activation of Antigen Presentation of cDC1 Cells.
J.Med.Chem., 67, 2024
|
|
8JGR
 
 | |
8JGT
 
 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
8JGP
 
 | |
8JGO
 
 | |
3NFS
 
 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
3NFP
 
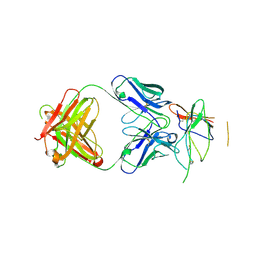 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
8ECE
 
 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Glu | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, L-asparaginase 2 | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
8ECD
 
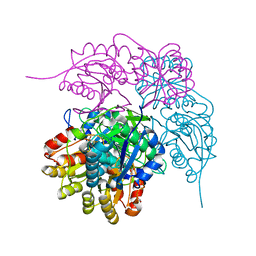 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Asp | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, CITRIC ACID, ... | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
4PZV
 
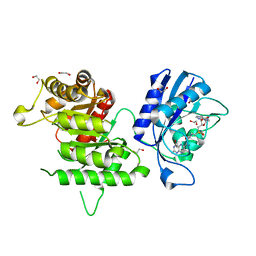 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with bisubstrate analog HPPK inhibitor J1D | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis.
Febs J., 281, 2014
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
3IU3
 
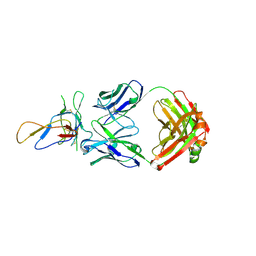 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
2YIQ
 
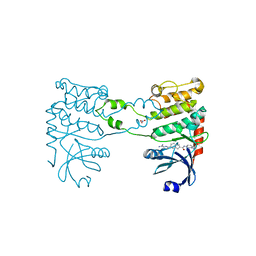 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1322 | | Descriptor: | (E)-5-(1-(2-CARBAMIMIDOYLHYDRAZONO)ETHYL)-N-(1H-INDOL-6-YL)-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
6J2E
 
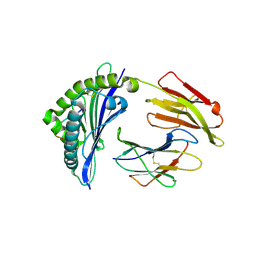 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP1 | | Descriptor: | Beta-2-microglobulin, EBOV-NP1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2G
 
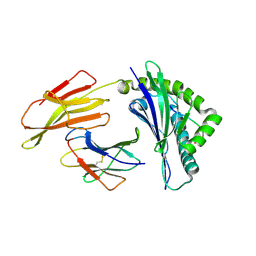 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP2 | | Descriptor: | Beta-2-microglobulin, EBOV-NP2, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6IX4
 
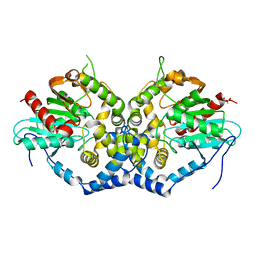 | | Structure of an epoxide hydrolase from Aspergillus usamii E001 (AuEH2) at 1.51 Angstroms resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hu, D, Hu, B.C, Hou, X.D, Wu, L, Rao, Y.J, Wu, M.C. | | Deposit date: | 2018-12-09 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Nearly perfect kinetic resolution of racemic o-nitrostyrene oxide by AuEH2, a microsomal epoxide hydrolase from Aspergillus usamii, with high enantio- and regio-selectivity.
Int.J.Biol.Macromol., 169, 2021
|
|
6J2D
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | HeV1, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
