3SFF
 
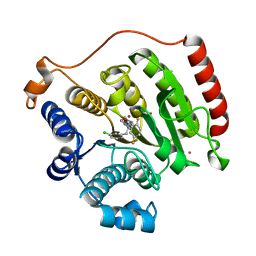 | | Crystal Structure of Human HDAC8 Inhibitor Complex, an Amino Acid Derived Inhibitor | | Descriptor: | (2R)-2-amino-3-(3-chlorophenyl)-1-[4-(2,5-difluorobenzoyl)piperazin-1-yl]propan-1-one, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
6KUJ
 
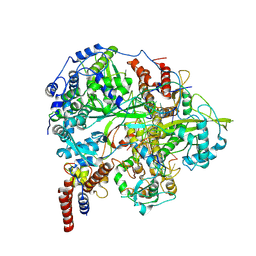 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 1 | | Descriptor: | 3'-cRNA promoter, 5'-cRNA promoter, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of influenza D virus polymerase bound to cRNA promoter in Mode A conformation
NAT NANOTECHNOL, 2019
|
|
8I4F
 
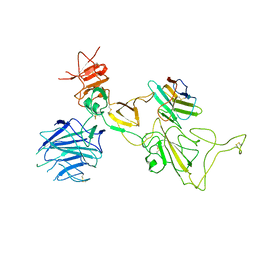 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
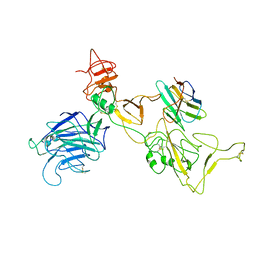 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4E
 
 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
9IXU
 
 | | Overall reconstruction of the Bax line | | Descriptor: | Apoptosis regulator BAX | | Authors: | Zhang, Y, Tian, L, Ge, X, Huang, G, Shi, Y. | | Deposit date: | 2024-07-29 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis of BAX pore formation.
Science, 388, 2025
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
8WHZ
 
 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHU
 
 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHW
 
 | | Spike Trimer of BA.2.86 with single RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHS
 
 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7FJP
 
 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7FJQ
 
 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2, SPERMINE | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
8HPU
 
 | |
8HQ7
 
 | |
