5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
1GCZ
 
 | | MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) COMPLEXED WITH INHIBITOR. | | Descriptor: | 7-HYDROXY-2-OXO-CHROMENE-3-CARBOXYLIC ACID ETHYL ESTER, CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, ... | | Authors: | Katayama, N, Kurihara, H. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
1GD0
 
 | | HUMAN MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) | | Descriptor: | CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, SULFATE ION | | Authors: | Kurihara, H, Katayama, N. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
3UIX
 
 | |
3UMW
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indazol-3-yl)methylene]-6-methoxy-7-(piperazin-1-ylmethyl)benzofuran-3(2H)-one | | Descriptor: | (2Z)-2-(1H-indazol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3(2H)-one, GLYCEROL, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational evolution of a novel type of potent and selective proviral integration site in Moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound.
J.Med.Chem., 55, 2012
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3VS2
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Parker, J.L, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VRZ
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS1
 
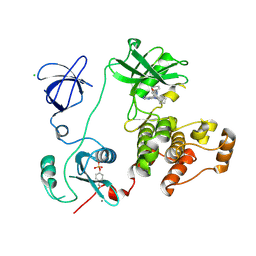 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS4
 
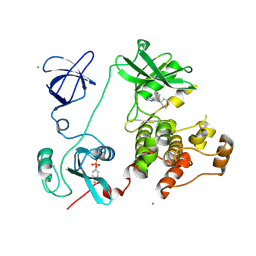 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS0
 
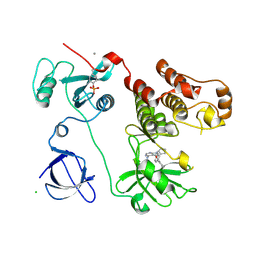 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS3
 
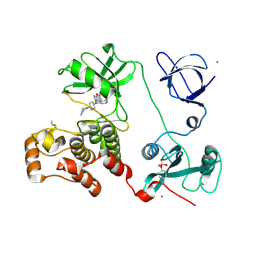 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
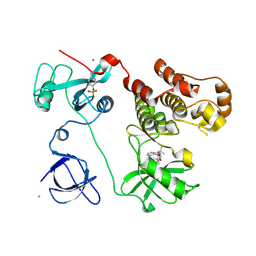 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
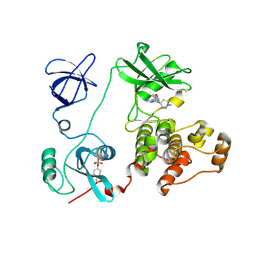 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VRY
 
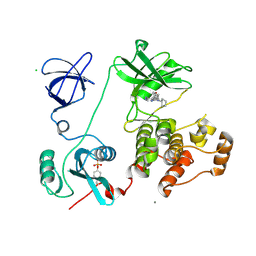 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS7
 
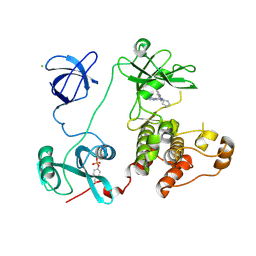 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Honda, K, Niwa, H, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
7W3L
 
 | | Crystal structure of LSD1 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE1
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE2
 
 | | Crystal structure of LSD2 in complex with trans-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE3
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, CITRATE ANION, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7C3G
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
